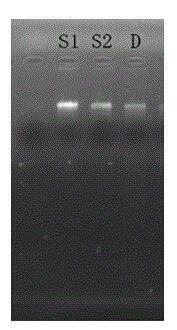
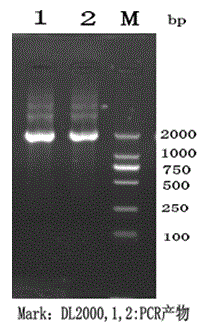
Paenibacillus polymyxa neopullulanase gene as well as method for producing neopullulanase through cloning and expression and fermentation
A technology of Bacillus polymyxa and new pullulanase, which is applied in the field of enzyme engineering, can solve the problems of less research on Bacillus polymyxa and has not yet realized industrialization, and achieve the effects of convenient cultivation, easy extraction, and improved stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] (1) Extraction of Bacillus polymyxa genomic DNA:
[0037] Cultivate Bacillus polymyxa in nutrient broth medium in a constant temperature and humidity incubator at 30°C overnight, take 0.5~2mL culture liquid (no more than 2×10 9 cells), centrifuge at 10,000 rpm for 30 seconds, discard as much supernatant as possible, and collect the bacteria.
[0038] Genomic DNA of the strain was extracted according to the bacterial DNA extraction kit (SK1201-UNIQ-10 Column Bacterial Genomic DNA Extraction Kit from Shanghai Sangon Bioengineering Technology Service Co., Ltd.).
[0039] Follow the steps below:
[0040] (1) Add 200μl buffer RB to resuspend the cells, centrifuge at 1000rpm for 30 seconds, discard the supernatant, resuspend the cells in 180μl buffer RB (Gram-negative bacteria) or 480μl 50mMNa2EDTA (pH8.0) (Gram Lambert-positive bacteria);
[0041] (2) Add 120μl lysozyme (20mg / ml in 10 mM Tris-HCl, pH 8.0) and mix well. Incubate at 37°C for 30-60 minutes. Centrifuge at 1...
Embodiment 2
[0097] Example 2: Escherichia coli-induced expression of the new pullulanase gene
[0098] (1) Extraction of PGEX 4T-1 plasmid
[0099] PGEX 4T-1 was extracted according to the instructions of the plasmid mini-extraction kit.
[0100] (2) Recovery of PCR products
[0101] Separate the target DNA fragment from other DNA as much as possible by agarose gel electrophoresis, then cut the agar block containing the target DNA fragment with a clean scalpel, and put it into a 1.5 mL centrifuge tube. (When determining the position of the DNA fragment, long-wavelength ultraviolet light should be used as much as possible, and the time of irradiation under ultraviolet light should be as short as possible).
[0102] (3) Digestion of vector and PCR product
[0103] The enzyme digestion reaction system is shown in Table 2:
[0104]
[0105] Digest overnight at 37°C (12-16 h), then inactivate at 75°C for 10 min.
[0106] (4) Purification of digested products
[0107] Cut the digeste...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com