Nucleic acid detection method based on surface plasmon resonance technology
A technology of surface plasmon and detection method, which is applied in the field of nucleic acid analysis, can solve problems such as difficult widespread application, pollution, and unstable structure, and achieve the effects of easy popularization and application, good biocompatibility, and sensitive detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
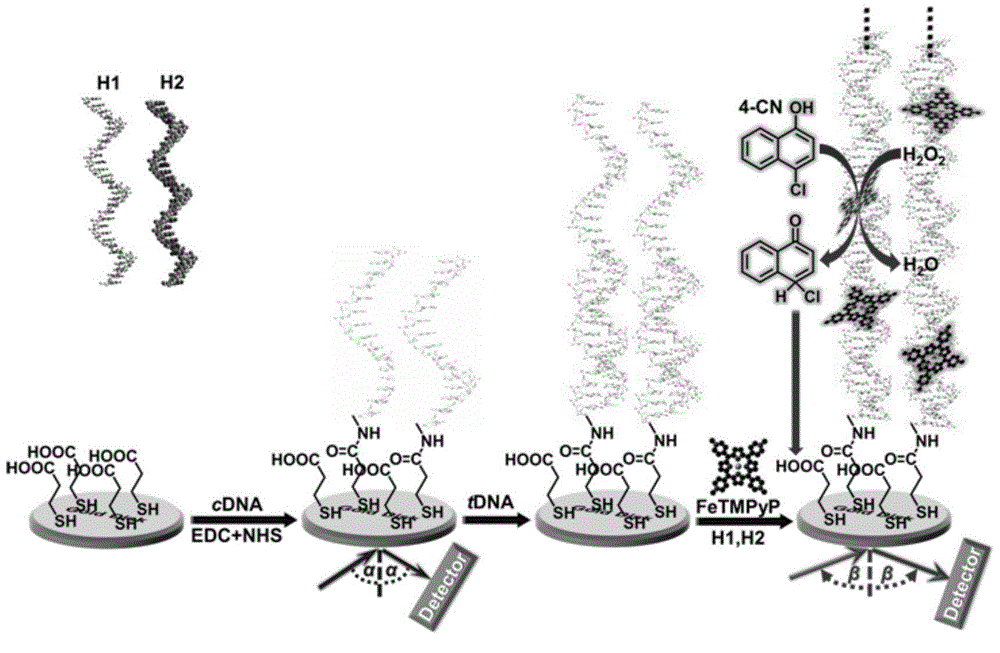
[0041] The SPR-based nucleic acid detection method uses a carboxylated SPR gold-plated chip as a carrier, EDC and NHS activate the carboxyl group, and then assembles the amino-terminated cDNA and ethanolamine onto the surface of the gold-plated chip through amidation reaction as a capture probe and seals the interface. The process is as follows:
[0042] (1) The bare SPR chip was prepared in piranha acid (concentrated H 2 SO 4 : Concentrated H 2 o 2 =1:3) After soaking for 10 minutes, then use the boiling mixture (concentrated ammonia water: concentrated H 2 o 2 : ultrapure water = 1:1:4) for 30 minutes, and finally washed twice with ultrapure water and ethanol 2 Blow dry; then, soak the treated gold flakes in 10-30mM MPA for 10-15 hours for pre-treatment, wash with ethanol and N 2 Blow dry and set aside.
[0043] (2) Soak the gold-plated sheet with (0.4M EDC / 0.1M NHS) for 30 minutes to activate the carboxyl groups on the surface of the gold sheet. Soak in 1-5 μM amino...
Embodiment 2
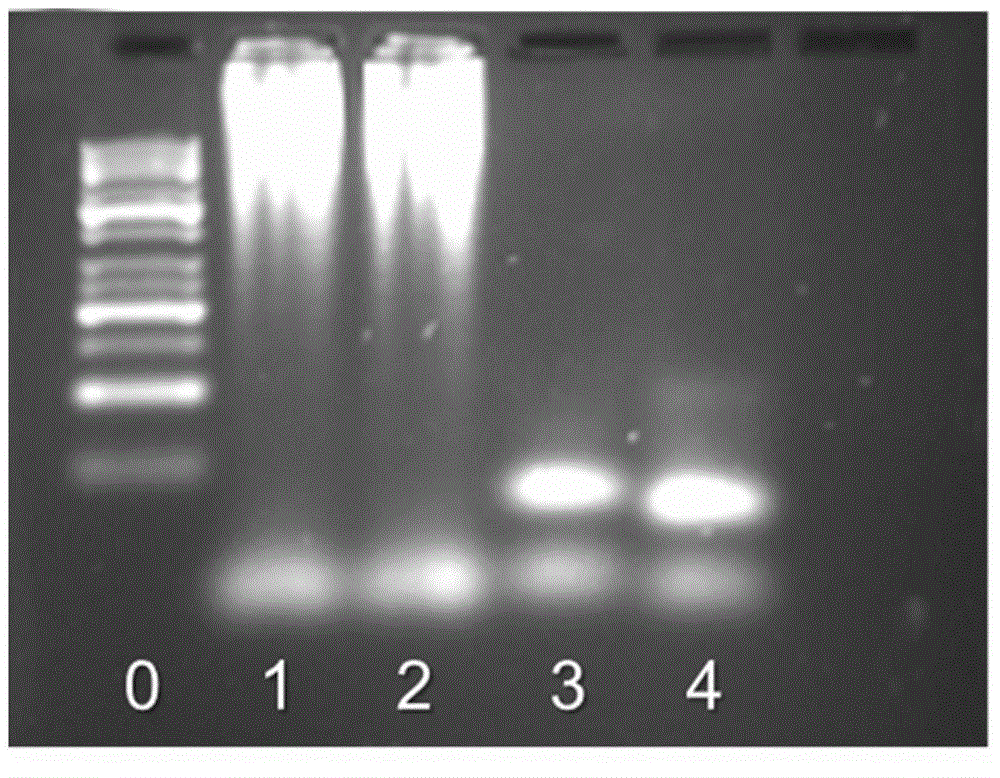
[0045] Generate long-chain dsDNA by hybridization chain cycle reaction, and verify its properties and structural characteristics:
[0046] Firstly, long-chain dsDNA is generated by hybridization chain cycle reaction, and the synthesis steps are as follows:
[0047] (1) Use buffer (0.5-1M dipotassium hydrogen phosphate and potassium dihydrogen phosphate to add 0.2-0.25M sodium chloride to adjust the pH to 7-7.5; the same below) to dissolve the cervical DNA (H1 and H2) solution.
[0048] (2) Dilute the neck loop DNA (H1, H2) to 10 μM, take 100 μL each and mix it, then quickly raise the temperature to 90-95°C, keep it for 5-10 minutes, take it out, and quickly cool it to room temperature in an ice-water bath to complete the hybrid chain cycle reaction to obtain long-chain dsDNA.
[0049] (3) Verify the HCR reaction product by agarose electrophoresis: prepare 2.5% agarose gel, add H1, H2 and HCR reaction product dsDNA into the electrophoresis tank respectively. The voltage was a...
Embodiment 3
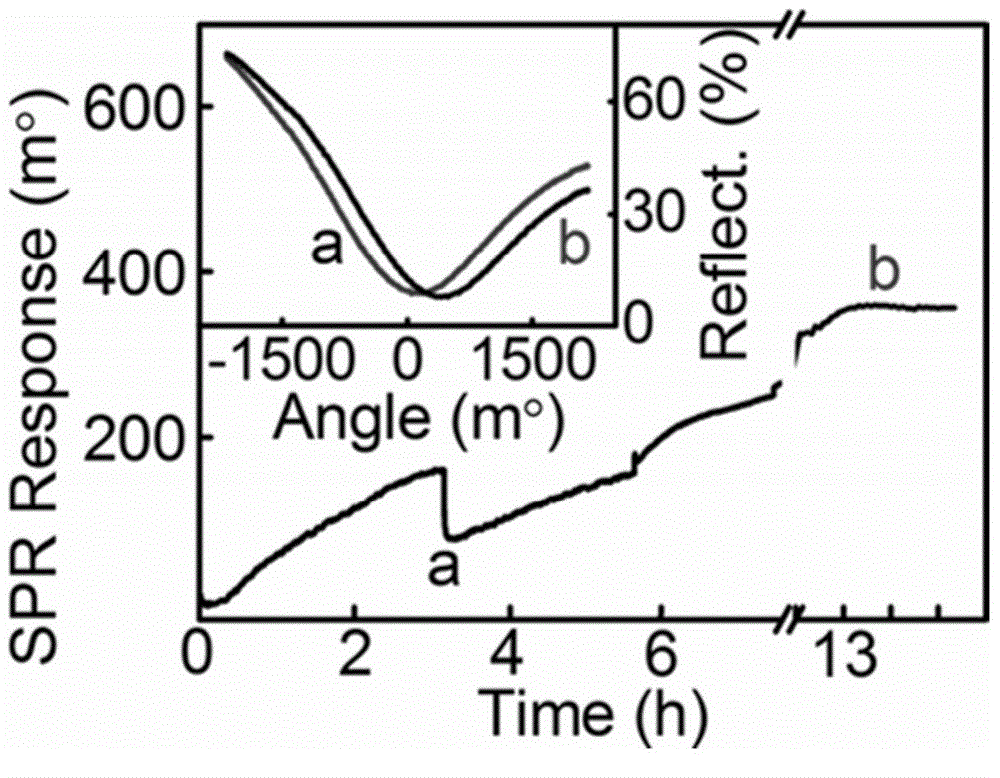
[0052] Using SPR technology to characterize the HCR reaction process, the reaction process is as follows:
[0053] (1) Rinse the surface of the Au / cDNA assembly layer with 100uL buffer solution, and obtain the baseline a after equilibration; add 1uM target DNA solution diluted with 100uL buffer solution, and react with the Au / cDNA assembly layer for 1-3 hours to obtain Au / cDNA / target DNA Layers were assembled and flushed with buffer to reach equilibrium baseline b.
[0054] (2) Dilute the neck loop DNA (H1, H2) to 5-10 μM with buffer solution, take 100 μL each and mix, then quickly raise the temperature to 90-95 °C, keep it for 5-10 minutes, take it out, cool it to room temperature in an ice-water bath, and then add it quickly In the Au / cDNA / target DNA assembly layer, react for 10-12 hours to obtain the Au / cDNA / dsDNA assembly layer, wash with buffer to reach the equilibrium baseline c.
[0055] Such as image 3 As shown, the results show that after adding the target DNA, bec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com