Method for preparing D-aromatic amino acid by cascade reaction
An aromatic amino acid and cascade reaction technology, applied in biochemical equipment and methods, enzymes, hydrolytic enzymes, etc., can solve the problems of low activity and loss, and achieve mild reaction conditions, high catalytic efficiency, and environmental friendliness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Cloning of D-carbamoylase gene
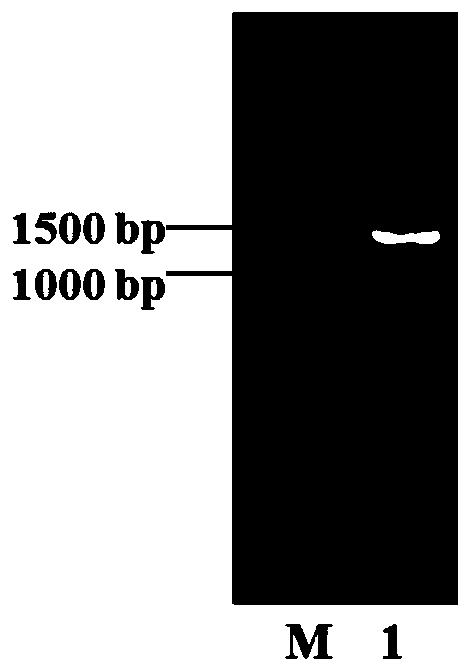
[0026] Cloning of D-N-carbamyl hydrolase gene from nitrate-reducing bacteria by one-step cloning method.
[0027] (1) First, the above-mentioned nitrate-reducing bacteria were cultured overnight at 37° C. using LB medium. After the bacterial cells were obtained by centrifugation, the total genomic DNA was obtained using a conventional bacterial genome extraction kit.
[0028] (2) Design primers according to the reported D-N-carbamyl hydrolase gene (upstream primer: CAGCAAATGGGTCGC GGATCC ATGACGCGGCGCATAAGG, SEQ ID NO.3; downstream primer: GTGGTGGTGGTGGTG CTCGAG TCACTCCACACCGGTCTGTGA, SEQ ID No.4), using the nitrate-reducing bacteria genome as a template for PCR, the system is as follows (μL): 10×PCR Mix 10, upstream primer 0.2, downstream primer 0.2, genome 0.2, DNA polymerase 0.2, ddH 2 O 9.2. The PCR program was: pre-denaturation at 95°C for 10 min, cleavage at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72...
Embodiment 2
[0029] Example 2: Construction and cultivation of recombinant Escherichia coli BL21(DE3) / pET28a-NihyuC
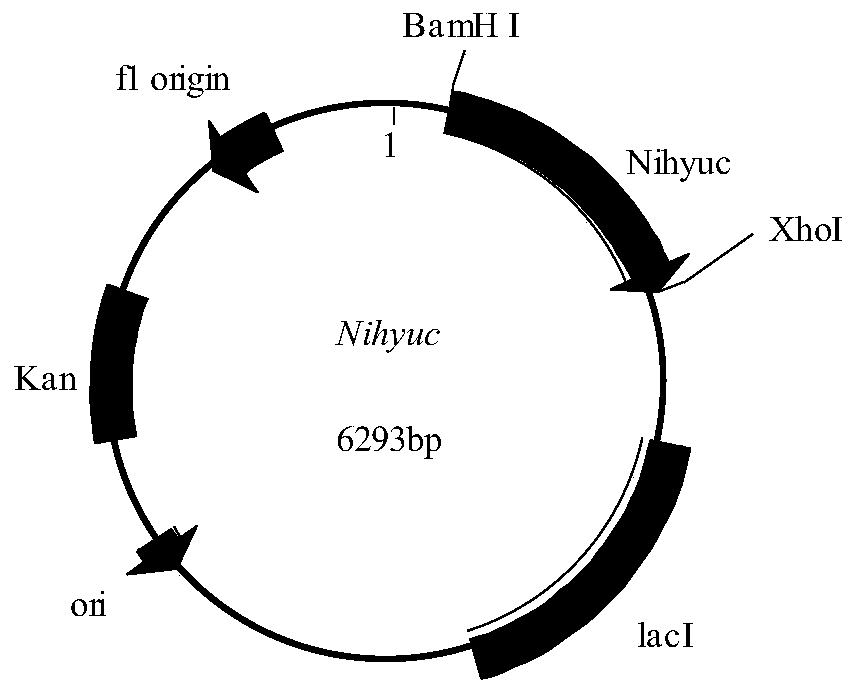
[0030]Plasmids pET28a and NihycC were digested with restriction endonucleases BamHI and XhoI for 3 hours in a water bath at 37°C. The next day, they were purified by agarose gel electrophoresis and the target fragments were recovered using an agarose recovery kit. At 37°C, the gene NihyuC was ligated with the digested plasmid pET28 using a one-step cloning method to obtain the recombinant expression vector pET28a-NihyuC( figure 2 ). Transform the constructed recombinant expression vector pET28a-NihyuC into Escherichia coli BL21(DE3) competent, coat a solid plate containing kanamycin-resistant LB, and perform colony PCR verification after overnight culture. The positive clone is the recombinant large intestine Bacillus BL21(DE3) / pET28a-NihyuC. Pick positive clones and culture them overnight in LB medium, then transfer them into fresh LB culture at 2% transfer amount the n...
Embodiment 3
[0031] Embodiment 3: Separation and purification of D-N-carbamyl hydrolase
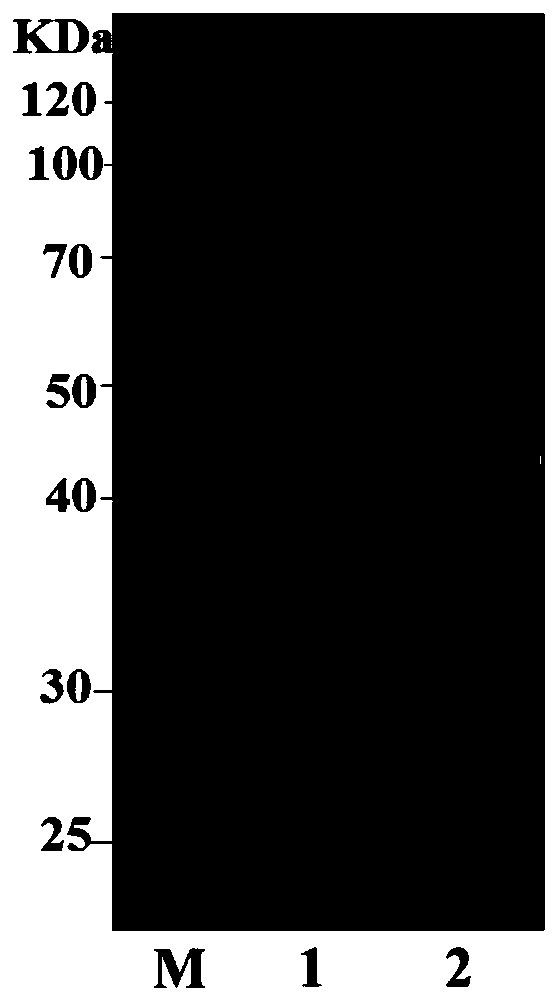
[0032] Suspend the recombinant cells in solution A (20mmol·L -1 Sodium phosphate, 500mmol·L -1 NaCl, 20mmol·L -1 imidazole, pH 7.4), the crude enzyme solution was obtained after sonication and centrifugation. The column used for purification is an affinity column HisTrap HP crude (nickel column), which is accomplished by using the histidine tag on the recombinant protein for affinity binding. First, use solution A to equilibrate the nickel column, load the crude enzyme solution, continue to use solution A to elute the breakthrough peak, and after equilibrium, use solution B (20mmol L -1 Sodium phosphate, 500mmol·L - 1 NaCl, 1000mmol·L -1 imidazole, pH 7.4) for gradient elution to elute the recombinant protein bound to the nickel column to obtain recombinant D-N-carbamoylase. Enzyme activity assay (DL-N-carbamoyl tryptophan as substrate) and SDS-PAGE analysis ( Figure 4 ). Depend on Figure ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com