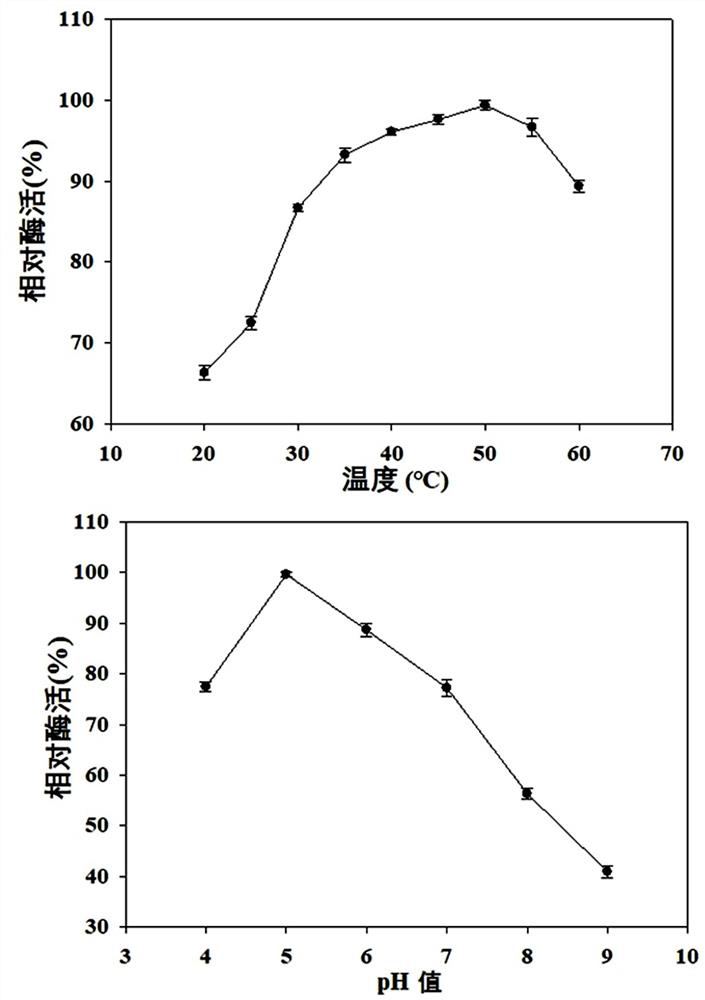
Alcohol dehydrogenase and its coding gene and its application in catalytic synthesis of (r)-phenylethylene glycol
A technology of phenylethylene glycol and alcohol dehydrogenase, applied in the field of enzyme engineering, can solve the problems of unsatisfactory, high cost, low selectivity of chemical synthesis method, etc., and achieve the effects of improving efficiency, promoting efficiency and reducing inhibition
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
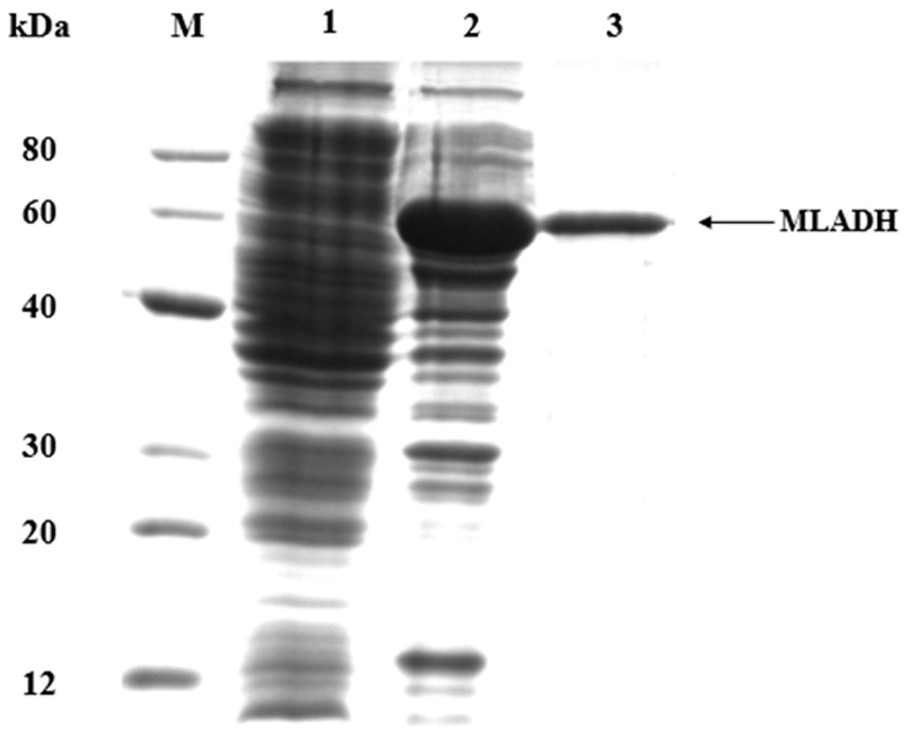
[0026] Example 1 Genome Mining of Microbacterium Alcohol Dehydrogenase and Construction of Recombinant Strains
[0027] Use Primer Premier software (Premier5.5, Palo Alto, CA, USA) to carry out primer design according to the oxidoreductase sequence deduced from the genome (GCF_000422405.1) of Microbacterium luticocti DSM 19459 registered in the NCBI database (as shown in Table 1 Show).
[0028] Table 1: Primer sequences
[0029] name Sequence (5'-3') name Sequence (5'-3') F1 ATGACNMGNACNGCNYTNATHACNG F7 ATGACNGGNGGNCARCCNCARTGGA R1 NCKNCCNCKNGCNGCNGCNCKNACN R7 NGCNARYTCNGCDATDATNGGRTGD F2 ATGGCNACNGAYWSNGCNYTNGAYG F8 ATGACNMGNYTNGGNMGNWSNGAYY R2 CCANSWNSWYTTNCKRTARTCYTTN R8 RTCYTGNGCNGCRTCNSWNGCNCKN F3 ATGGCNGGNACNWSNGGNCAYGTNA F9 ATGWSNMGNTTYACNGGNAARGTNG R3 YTGNACNSWCCANCCNCCRTCNSWN R9 RTGNGCNSWRTANCCNCCRTCNACN F4 ATGACNGAYGCNMGNAAYGAYGAYG F10 ATGCCNTAYMGNGTNATHGTNACNG R4 NGCNGGNGCRTCDATNCCD...
Embodiment 2
[0039] The construction of embodiment 2 double enzyme coupling catalytic system
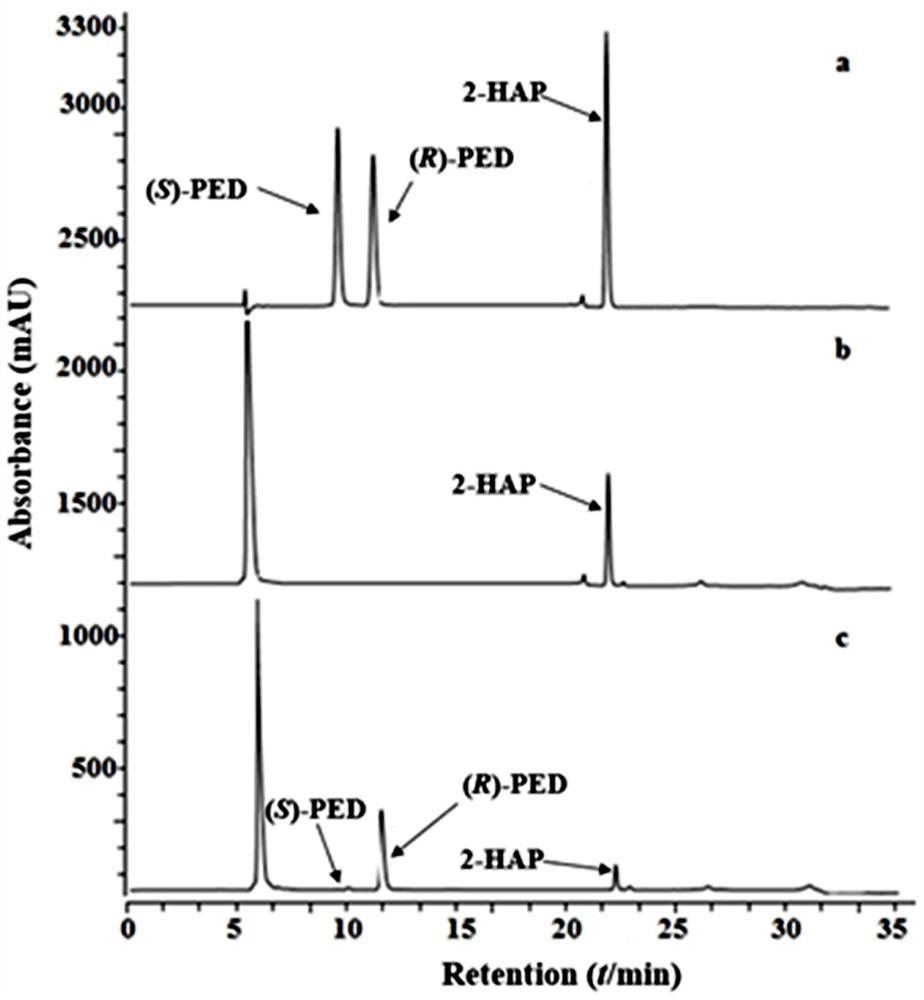
[0040] The recombinant protein FDH from Candida parapsilosis was passed through the purification system ( GE Healthcare), the affinity column was filled with prepacked Ni Sepharose TM (HisTrap HP; GE Healthcare, Piscataway, NJ, USA). Formate dehydrogenase FDH activity was determined by measuring the increase in absorbance at 340 nm. The reaction mixture (0.1 mL) for the enzyme assay included 0.1 M sodium formate, 1 mM NAD+, 0.1 M KHPO 4 buffer (pH 7.5) and the appropriate enzyme. The specific activity of the enzyme is 140U / mg. Purified MLADH and FDH (100 μL each) were added to a potassium phosphate buffer reaction system (1 mL) containing 1.0 g / L 2-HAP, 1.0 g / L sodium formate and 2 mM NAD+. The reaction system was transformed at 37°C and 200r / min for 2h. After the reaction was completed, the reaction solution was centrifuged, and 100 μL of the supernatant was dissolved in 900 μL of methanol ...
Embodiment 3
[0044] The optimization of embodiment 3 two-phase catalytic system
[0045] Firstly, the biphasic catalytic system of dual enzyme reaction was optimized. The reaction mixture (10 mL) consisted of 0.1 M potassium phosphate buffer (pH 7.0) containing 0.5 g / L sodium formate, 2 mM NAD+, two enzymes (MLADH:FDH=1:10) and an organic solvent (phthalic acid Dibutyl ester, ethyl caprate, ethyl laurate, n-heptane, ethyl acetate, n-hexanol, n-hexane or soybean oil), the concentration of 2-HAP is 5g / L, and the ratio of aqueous phase to organic phase is 1:1. The mixture was stirred at 37 °C for 2 h. Sample 100 µL of each organic layer and dissolve in 900 µL methanol. Product content was determined by HPLC. In addition, the biotransformation of 2-HAP by the dual enzymes was also analyzed using different ratios of aqueous and organic phases (1:0, 4:1, 3:1, 2.3:1, 1.5:1 and 1:1).
[0046] Secondly, the optimal initial concentration of the substrate 2-HAP in the catalytic system was explor...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com