Novel method for detecting enterobacter sakazakii, application and detection kit
A technology of Enterobacter sakazakii and detection kit, applied in the field of food-borne pathogen detection, can solve the problems of undiscovered patent publications, etc., achieve easy standardization and automatic operation, high reliability and sensitivity, and reduce detection Effects of time and false positive results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
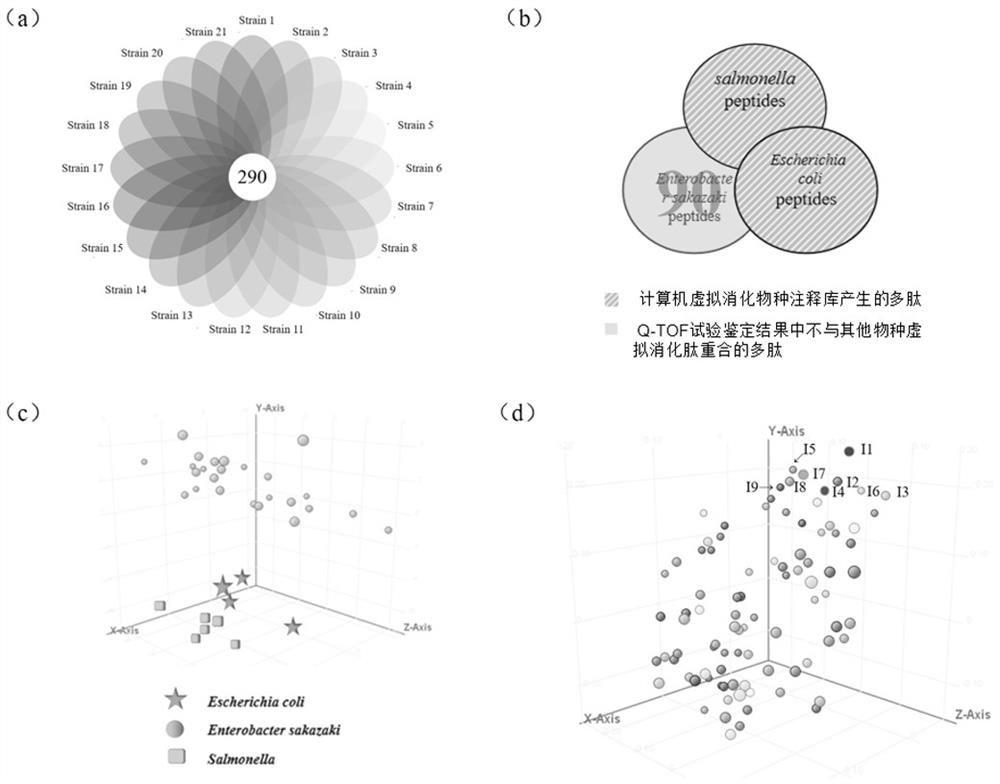
[0055] Example 1: Mining of characteristic peptides
[0056] 1) Non-targeted proteomic analysis: UPLC-Q Exactive was used to conduct non-targeted proteomic identification analysis on 21 strains of Enterobacter sakazakii, and then the data was processed by Mascot. Liquid chromatography conditions: flow rate 0.2 mL / mL; injection volume 10 μL; elution program: 0~5 min, keep phase B from 5%; 5~30 min, increase phase B from 5% to 40%; 30~ For 40 min, 95% of phase B was maintained. Mass spectrometry conditions: heated electrospray ion source (HESI); positive ion mode; full scan mode, particle range: m / z 300-1500; resolution 70 000; maximum capacity of Ctrap (AGC target): 100000; maximum injection time of Ctrap : 100 ms; sheath gas velocity: 40 arb; auxiliary gas flow rate: 3 arb; spray voltage: 3.5 kV; capillary temperature: 300 ℃. Mascot software identification conditions: select SwissProt for protein database, trypsin for protease type, 1 maximum missed cleavage site, Carbamidom...
Embodiment 2
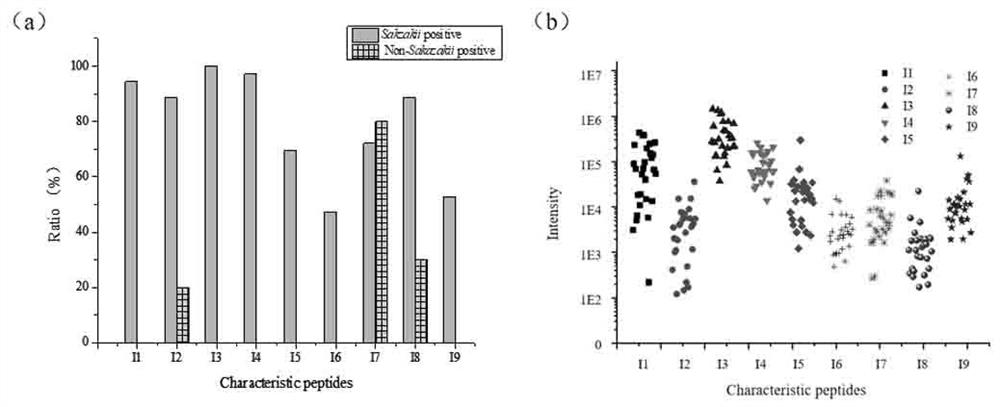
[0059] Example 2: Specific verification and optimization of candidate characteristic peptides
[0060] 4) Standard strains of Enterobacter sakazakii, Salmonella and Escherichia coli were cultured in nutrient agar medium (1% peptone, 0.5% NaCl, 0.3% beef extract, 2% agar) respectively, and cultured at 37 °C for 24 h.
[0061] 2) Bacteria collection and sample pretreatment: add 0.5 ml Buffer 1 (C) and (1) treated bacteria liquid, mix well, and ultrasonically break for 10 min. Then, add 100 μL of Buffer 2 (D) and mix quickly, and react in the dark for 15 minutes. Then 4 μL of trypsin (E) was added and digested in a water bath at 50 °C for 15 min. Add 0.5 μL formic acid (F) to terminate trypsin digestion and analyze samples directly or store at -20 °C until analysis.
[0062] 5) MRM pattern targeted analysis of LC-DSI / QQQ. Chromatographic conditions: Vydax C18 column (2.1mm×150mm, 5 μm); flow rate 0.3 ml / min; injection volume 2 μL; mobile phase A is an aqueous solution containi...
Embodiment 3
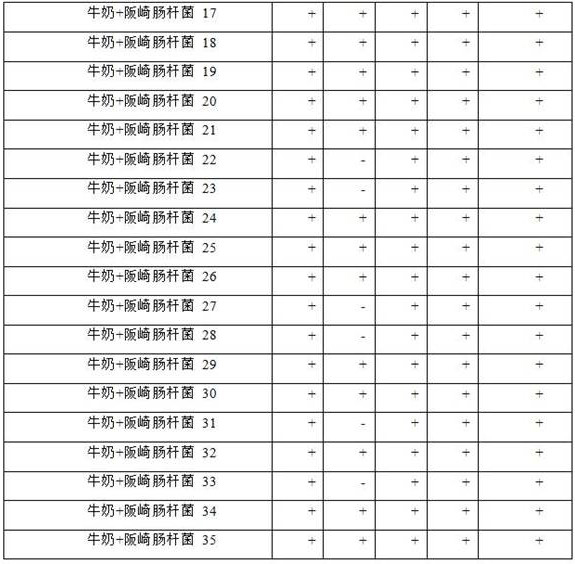
[0072] Example 3: Application of characteristic peptides in the detection of Enterobacter sakazakii contamination in milk powder samples
[0073] The target pathogenic bacteria liquid was added to pure milk powder (Yili, China) to a concentration of 100 CEU / mL. Take 0.1 mL contaminated milk powder sample, add it to 0.9 mL LB medium, and incubate at 37°C for 12 h. Subsequent steps are consistent with the processing methods (2)-(4) in Example 2. The evaluation results are shown in Table 3.
[0074]
[0075]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com