Acetonic acid oxidase gene, recombinant expression plasmid and transformation strains thereof
A pyruvate oxidase and expression plasmid technology, which is applied in the fields of oxidoreductase, genetic engineering, plant gene improvement, etc., can solve the problems of gene leakage expression downstream of the Lac promoter, reduce fermentation costs, facilitate industrial production, The effect of reducing the cost of enzymes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 , AvPyOD gene cloning
[0038] First design the following primer pairs:
[0039] Upstream: 5'-CATG CCATGG GAATGCATCACCATCACCATCATCACTCAGATAACAAAATTAACATC-3';
[0040] (Nco I restriction site)
[0041] (simultaneously add the nucleotide sequence corresponding to 9 amino acid residues MGMHHHHHH for purification)
[0042] Downstream: 5'-CGC GGATCC TTATCATTTGATGTATTTAGATTCTAAGCCTTCAGC-3';
[0043] (BamHI restriction site)
[0044] The AvPyOD gene was amplified by PCR using the genome of Aerococcus viridans ATCC10400 as a template and using the above primer pairs.
[0045] PCR system:
[0046] 5μl 10×PCR reaction buffer, 1μl downstream primer (50pM), 1μl downstream primer (50pM), 2μl Coccus viridans ATCC10400 genomic DNA (50ng / μl), 5μl dNTP mix (10mM, each 2.5mM), 0.5μl Pyrobest DNA polymerase (5U / μl), sterilized deionized water to 50μl.
[0047] PCR conditions:
[0048] 95°C for 5 minutes, then 30 cycles (each cycle condition is 95°...
Embodiment 2
[0050] Example 2 , Construction of recombinant expression plasmid pSMLPyOD
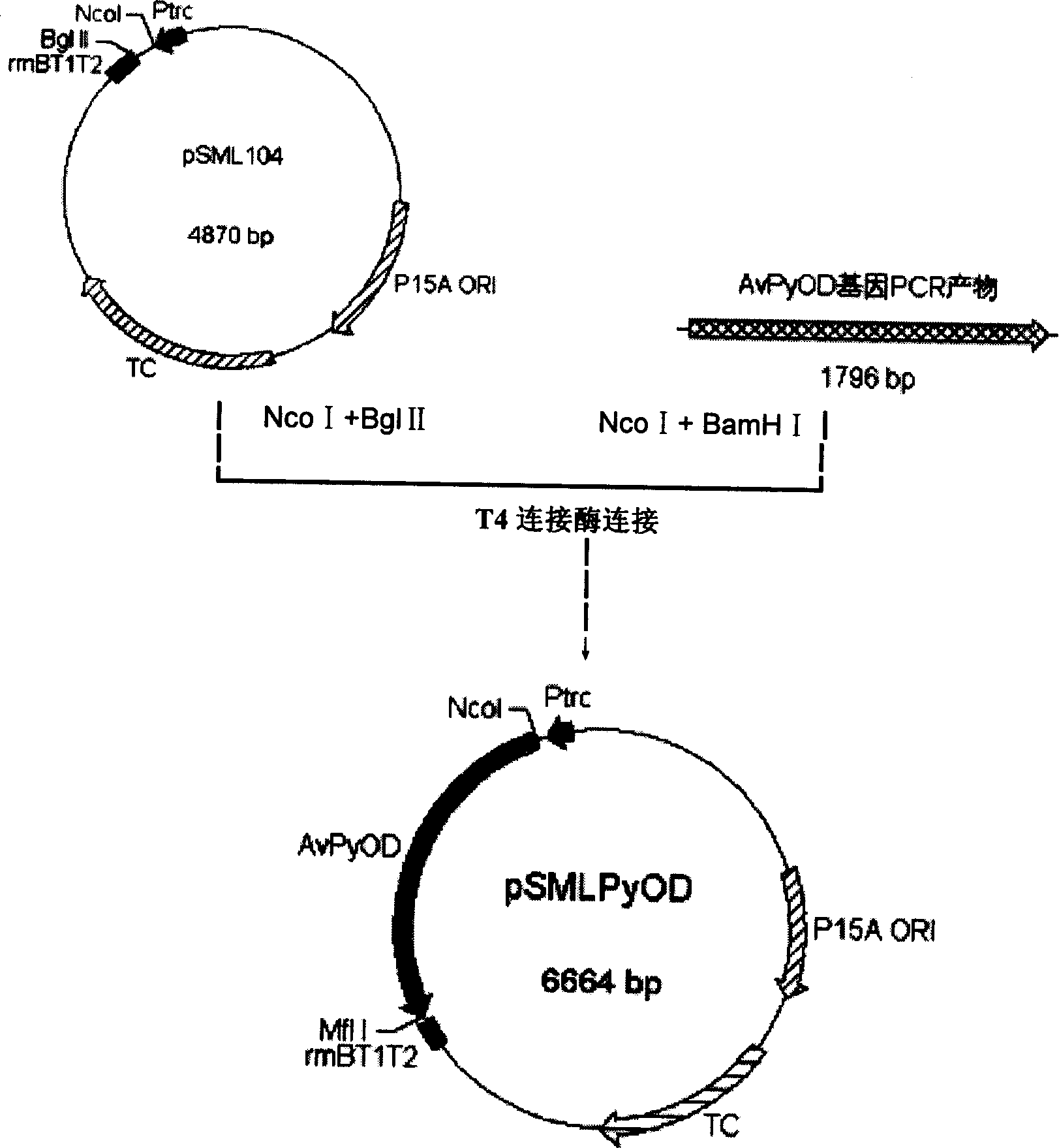
[0051] The plasmid pSML104 used in this example was obtained according to the method in the literature (Li Y, et al. Eur. J. Biochem., 1999, 262: 713-719). This plasmid uses trc as a promoter and has its own expression element , the transcription terminator is rrnb, the replicon is p15a (middle copy), and contains a tetracycline (Tc) resistance gene.
[0052] The amplified product obtained in Example 1 was double-digested with restriction endonucleases Nco I and BamH I to obtain the AvPyOD gene with cohesive ends at both ends; pSML104 plasmid; then connect the two digested fragments, the connection conditions are: 2.5 μl 10×T4 DNA ligase buffer, 2U T4 DNA ligase, sterilized deionized water to 25 μl, 16 ° C ligation reaction for 16 hours; the product obtained after ligation is For the recombinant expression plasmid pSMLPyOD; the whole construction process is as follows figure 1 shown.
[0053] In ...
Embodiment 3
[0060] Example 3 , the acquisition of genetically engineered strain DH5α / pSMLPyOD
[0061] Transform the recombinant expression plasmid pSMLPyOD into Escherichia coli DH5α (Dalian Bao Biological Company) {genetic characteristics are F-, supE44, lacU169, φ80d / lacZ In M15, hsdR17, recA1, endA1, gyrA96, thi-1, relA1}, the obtained transformant DH5α / pSMLPyOD is as follows:
[0062] Take 1 μl of the recombinant expression plasmid pSMLPyOD, add it to 100 μl of Escherichia coli DH5α competent cells, bathe in ice for 20 minutes, bathe in water at 42°C for 90 seconds, and bathe in ice for 3 minutes, add LB medium (peptone 1%, yeast extract 0.5%, chlorine NaCl 1%) 900 μl, incubated at 37°C for 1 hour; then 100 μl was spread on a selective plate containing tetracycline (sodium pyruvate 10g peptone 10g, yeast extract 5g, sodium chloride 10g magnesium sulfate 0.5g horseradish over Oxidase 500U anisidine 10mg tetracycline 50mg agar powder 15g deionized water to 1000ml), incubate at 30...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com