High-efficiency preparation method of beta-amylase
An amylase and seed technology, applied in the field of enzyme engineering and bioengineering, can solve the problems of low beta-amylase activity, poor heat resistance and high cost, and achieve the effects of high enzyme activity level, good thermal stability and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] The sweet potato RNA that embodiment 1 Trizol method extracts
[0059] (1) Clean the sweet potato, cut it into small pieces, grind it into powder in liquid nitrogen, transfer about 50~100 mg sample into a 1.5 mL centrifuge tube, add 1 mL Trizol reagent, invert and mix well to suspend the sample in the reagent middle.
[0060] (2) After the suspension was left at room temperature for 5 minutes, 0.2 mL of chloroform was added, the centrifuge tube was shaken vigorously for 15 seconds, left at room temperature for 5 minutes, and then centrifuged at 12,000 rpm for 10 minutes at 4°C.
[0061] (3) Carefully pipette the upper aqueous phase into a new centrifuge tube, add 0.5 mL of isopropanol, mix well, place at room temperature for 10 min, and then centrifuge at 12,000 rpm for 10 min at 4°C.
[0062] (4) Discard the supernatant, add 1 mL of 75% ethanol to wash the precipitate, and centrifuge at 12,000 rpm for 5 min at 4°C.
[0063] (5) Discard the supernatant, and dry the pe...
Embodiment 2
[0064] Example 2 Synthesis of cDNA and amplification of β-amylase gene
[0065] (1) Synthesis of cDNA by reverse transcription
[0066] Reaction system (20 μL): 1 μg RNA, reverse transcription primer 10 mM oligdT-18 1 μL, 5×Buffer 4 μL, 10M dNTPs 1 μL, M-MLV reverse transcriptase 1 μL, remaining volume with RNase-free water make up.
[0067] Reaction conditions: first add RNA and reverse transcription primers to the reaction tube, make up the volume to 10 μL with RNase-free water, 70°C, 5 min, and place on ice for 5 min; then add the remaining reagents, make up the volume to 20 μL with RNase-free water, 42°C, 60 min.
[0068] (2) PCR amplification containing the complete β-amylase gene BBA, the primers used are:
[0069] BBA-F: 5′-a agatct ccatggctccaatccccggt-3′
[0070] BBA-R: 5′-a agatct gctcctccttcaccttcag-3′
[0071] Amplified using Takara's ExTaq DNA polymerase
[0072] reaction system:
[0073] Volume (μL) Sterilized double distilled water 33...
Embodiment 3
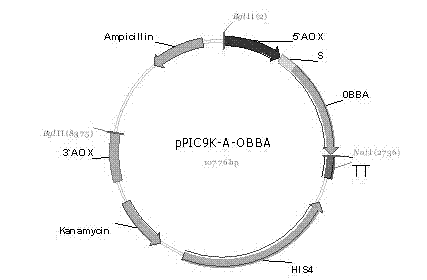
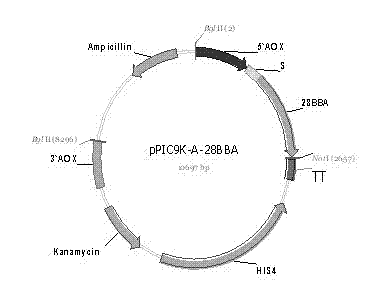
[0076] Example 3 Construction of Secreted Expression Vector pPIC9K-(A / B / C / D)-OBBA
[0077] Using 10 pg of the Teasy-BBA plasmid as a template, PCR amplifies the target fragment 0BBA, and the primers used are:
[0078] 0BBA-F: 5′-atggctccaatccccggt-3′,
[0079] 0BBA-R: 5'-tcaatcaaacgggtttgagccatc-3';
[0080] Reaction system and reaction condition are the same as embodiment 2.
[0081] The amplified product was the 1500 bp open reading frame sequence of the β-amylase gene except the signal peptide coding region.
[0082] The target gene OBBA and Pichia pastoris secreted expression vector pPIC9K-A were respectively used Bgl II and Bam HI was subjected to double enzyme digestion, and reacted overnight at 37°C; after the plasmid digestion product and PCR product were purified with a purification kit, T 4 DNA ligase was ligated at 4°C for 14 h. Transform the ligation product into Escherichia coli JM109, smear the LB plate containing 50 μg / mL kanamycin, select positive clon...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com