Engineering bacterium for producing Phospholipase A2 (PLA2) and applications thereof
A technology for producing phospholipids and engineering bacteria, applied in the field of bioengineering, can solve the problems of low stability of plasmids, insufficient stability of recombinant strains, cumbersome operation, etc., and achieve the effect of separation and purification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
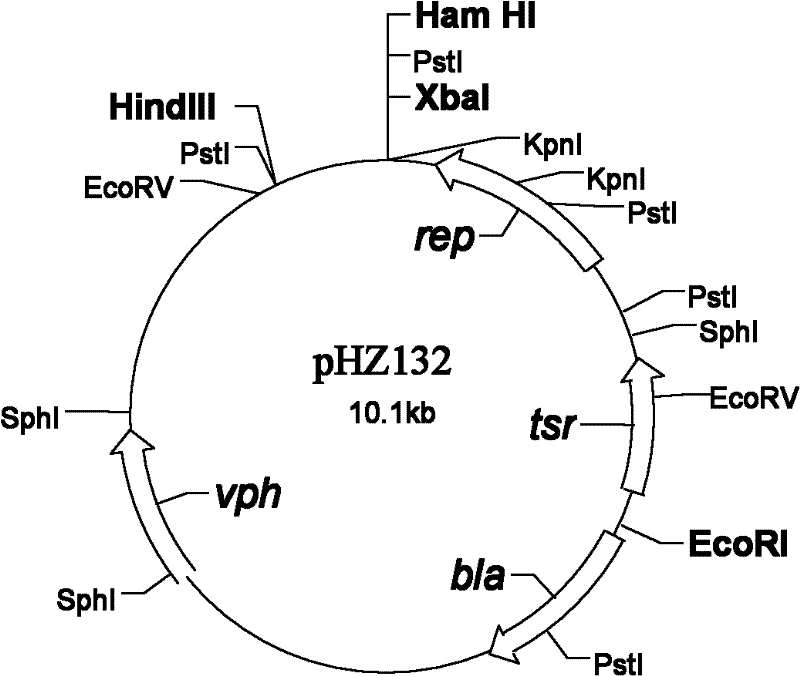
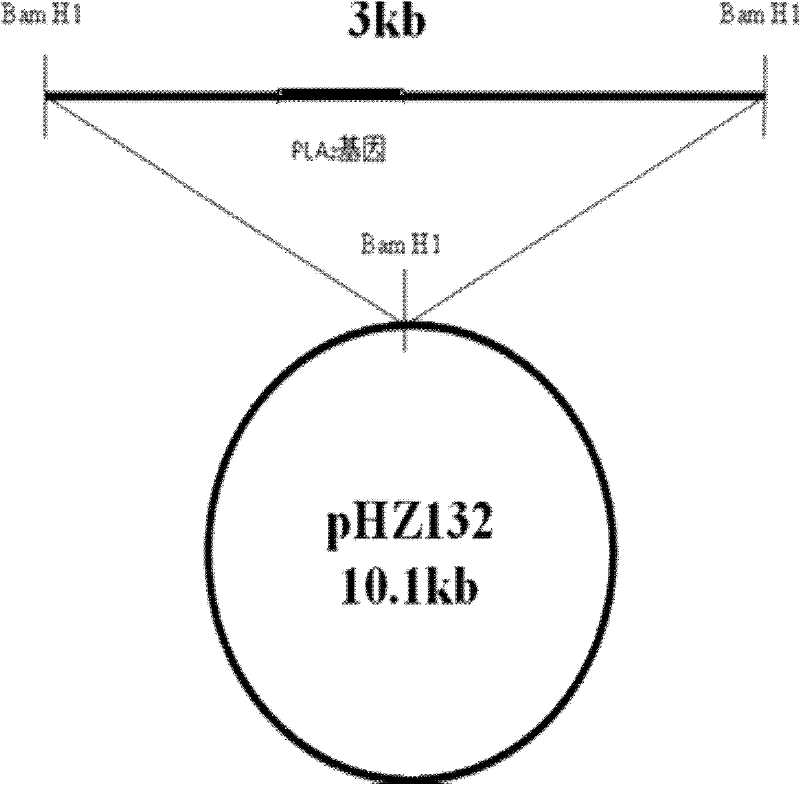
[0073] A phospholipase A 2 The preparation method of gene and leader signal peptide plasmid, its step is:
[0074] A. Used to amplify phospholipase A 2 Synthesis of primers for genes:
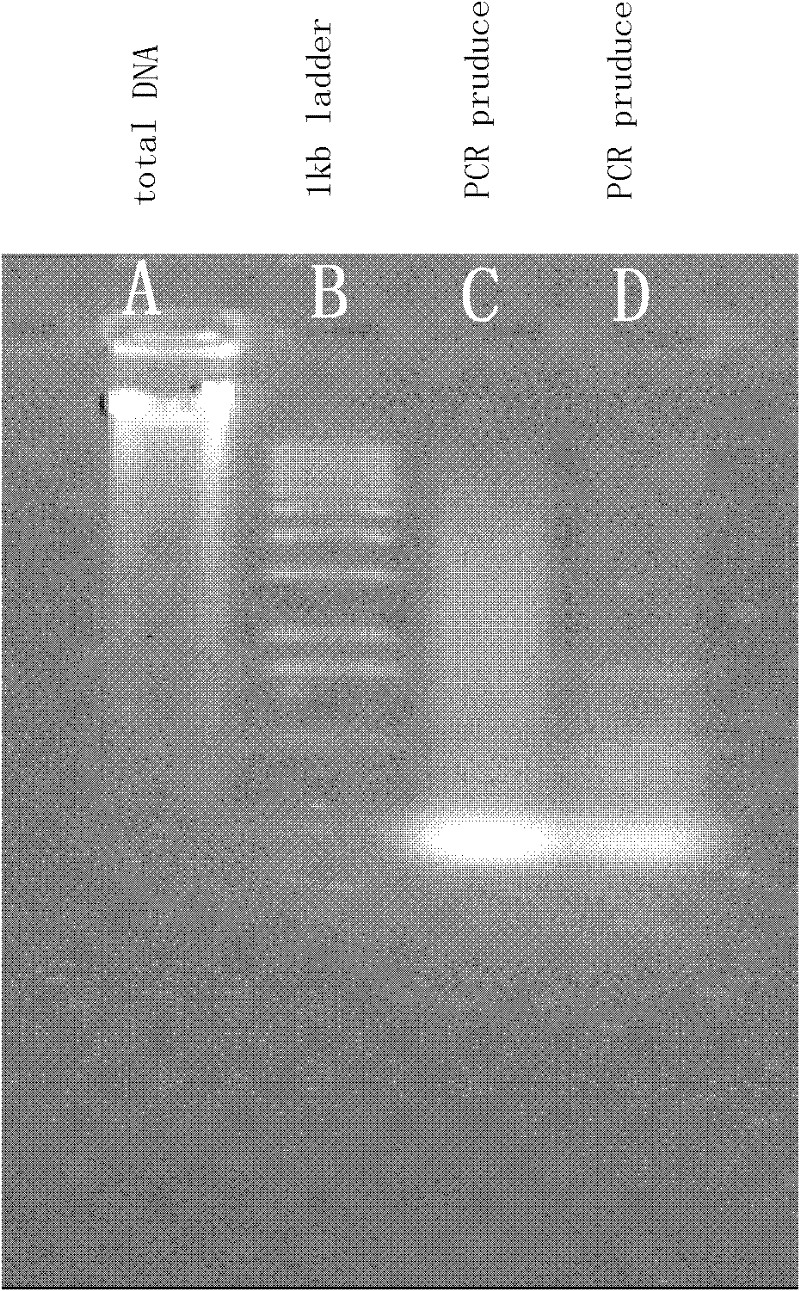
[0075] Access http: / / www.ncbi.nlm.nih.gov / GenBank, based on Streptomyces coelicolor A3(2) phospholipase A 2 Characterization of the DNA sequence from which Streptomyces phospholipase A 2 gene, and accordingly designed two PCR primers Oligo1 (SEQ ID NO.03) and Oligo2 (SEQ ID NO.04) with a length of 21 base pairs, and the designed primers were synthesized by a biotechnology company. The sequence and properties are detailed in the table below.
[0076] The characteristics of table 2 primers D11 and D12
[0077]
[0078] B, extraction of Streptomyces violaceum 2917 total DNA:
[0079] Dissolve 500ml of S.violaceoruber 2917 mycelia in 10ml of 2mg / ml LRTE solution (25mmol / LTris-HCl, pH8.0, 100mmol / L EDTA, RNase 50ug / ml.), lyse at 37°C until the solution is clear and translucent ;Add proteas...
Embodiment 2
[0098] a phospholipase A 2 The preparation method of engineering bacterium, its step is:
[0099] A. Add 25mL YEME medium to a 250ml Erlenmeyer flask with a stainless steel spring, and add glycine to a final concentration of 0.5%;
[0100] B. Inoculate 0.1 mL of Streptomyces S.lividans 1326 spore suspension, culture in a shake flask at 30°C for 36-40 hours; collect the bacteria by centrifugation;
[0101] C. Wash twice with 10.3% (W / V) sucrose solution; add 5 mL of lysozyme solution to the mycelium precipitate, and enzymolyze it in a water bath at 30° C. for 30-60 minutes;
[0102] D. Blow and suck 4 times with a sterile pipette, and continue to incubate in a water bath at 30°C for 15 minutes; add 5mL P buffer, blow and suck again; transfer to a filter equipped with absorbent cotton and filter; centrifuge the filtrate at 3,000rpm for 10 minutes to obtain protoplast precipitation;
[0103] E. Wash the protoplast pellet with 5mL P buffer solution for 1-2 times, and finally su...
Embodiment 3
[0106] The enzyme production ability comparison of engineering strain Streptomyces lividans LH001 and other bacterial strains etc., its steps are:
[0107] A. Inoculate the engineering bacteria Streptomyces lividans LH 001, the starting bacterium Streptomyces lividans 2917, the host strain Streptomyces lividans lividans 1326, the recombinant strain Streptomyces lividans lividans LEE2265, etc. into 100mL YEME medium respectively, and cultivate them at 30°C for 72 hour, 5000 rpm / centrifugation for 10 minutes, and the supernatant was obtained as the crude enzyme solution.
[0108] B. Qualitative and quantitative determination of phospholipase A by gas chromatography 2 vitality:
[0109] Principle: Phospholipase A 2 It can catalyze the hydrolysis reaction of the C-2 ester bond of glycerophospholipids to generate lysophospholipids and fatty acids, and qualitatively and quantitatively determine phospholipase A by measuring the types and quantities of fatty acids generated by the e...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com