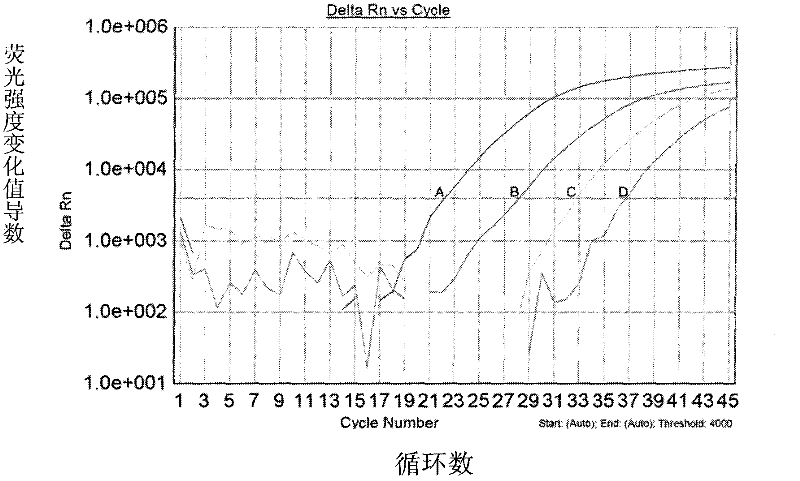
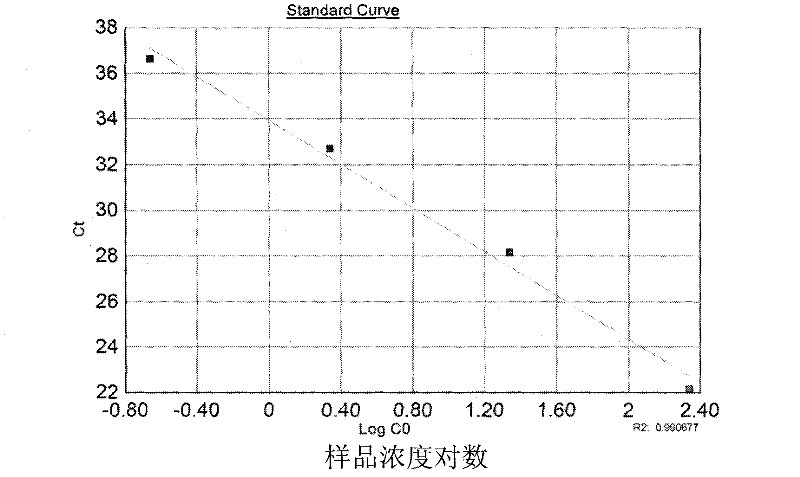
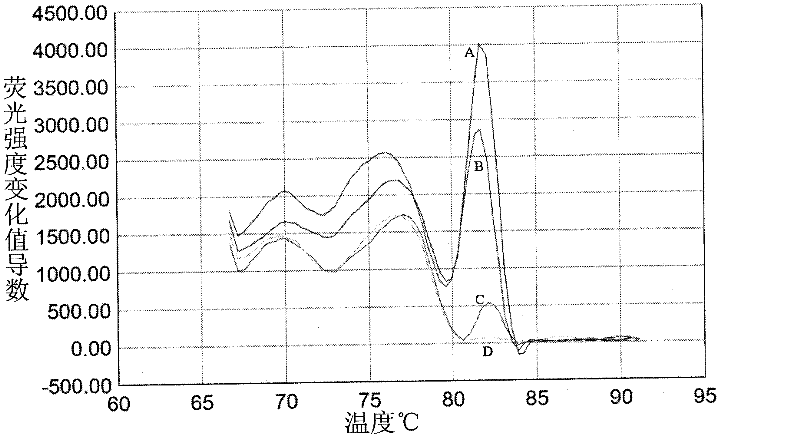
Method for amplifying and genotyping nucleic acid genes of human papilloma virus and assay kit for same
A viral nucleic acid and gene amplification technology, which is applied in the field of human papillomavirus nucleic acid gene amplification and typing, can solve the problems of high cost, cross-hybridization risk, and only typing, etc., and achieves a high degree of automation and eliminates Effects of Nucleic Acid Cross Contamination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Preparation of primers and probes
[0041] Primer (primer sequence: SEQ.ID.NO: 1-26) and probe (quantitative probe sequence: SEQ.ID.NO: 27-39; typing probe sequence: SEQ.ID.NO: 40-41) All were synthesized by an automatic nucleic acid synthesizer, and were synthesized by Shanghai Sangon Biotechnology Co., Ltd. in this example.
[0042] Among them, the 5' end of the quantitative probe is labeled with a fluorescent luminescent group FAM (emission wavelength: 522nm), and the 3' end is modified with a BHQ-2 fluorescence quenching group.
[0043] The 5' end of the typing probe is labeled with fluorescent groups HEX (emission wavelength: 555nm) and ROX (emission wavelength: 608nm), and the 3' end is modified with BHQ-2 fluorescence quenching group.
[0044] The above primers and probes were all dissolved in double-distilled water and made into a 4 μM solution for later use.
Embodiment 2
[0045] Example 2 Sample Collection
[0046] Use a special cervical brush for sampling. The central bristle part of the cervical brush is gently inserted into the cervical canal, so that the shorter bristles can fully touch the cervix, gently press the cervical brush forward, and rotate in the same clock direction Cervical brush for 3-5 weeks, scrape cervical cells without leaving a dead angle, and stay for 10 seconds. The sampling range included the entire transformation zone (Transformation Zone, that is, the junction area between cervical squamous epithelium and cervical columnar epithelium).
[0047] Put the cervical brush that has collected the sample into the sample preservation tube (contains 1ml of sample preservation solution, which can make the DNA intact and inhibit the growth of bacteria), cover it tightly and make a label (indicate the name and date).
[0048] Sample preservation solution formula: phosphate buffer solution 10mM, NaCl 138mM, KCl 2.7mM, 0.05% NaN3, ...
Embodiment 3
[0049] Example 3 Extraction of viral DNA
[0050] The DNA of HPV virus was extracted by magnetic bead method automatic nucleic acid extractor (or silica gel column method). Its working principle and steps include cracking, adsorption, cleaning, elution and recovery.
[0051] The buffer formulations are as follows:
[0052] Lysis buffer formulation: 0.5mg / ml proteinase K, 0.01M Tris-HCl (trishydroxymethylaminomethane), 0.001M EDTA (ethylenediaminetetraacetic acid), pH=8.0.
[0053] Adsorption buffer formulation: 5.5M GuSCN (guanidine isothiocyanate), 20mM EDTA, 10mM Tris-HCl, 65mM dithiothreitol, 40g / L silica-based magnetic beads, pH=6.5.
[0054] Rinse solution I formula: 5.5M GuSCN (guanidine isothiocyanate), 20mM EDTA, 10mM Tris-HCl, 65mM dithiothreitol, pH=6.5.
[0055] Rinse solution II formula: 25% (v / v) isopropanol, 25% (v / v) ethanol (96%), 50% double distilled water, 0.1M NaCl.
[0056] Eluent formula: 10mM Tris-HCl, pH=8.0, or double distilled water. Finally, 30-5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com