Carrier capable of showing and expressing helicobacter pylori protein on surface of lactococcus lactis, and preparation method and application of carrier
A technology of Helicobacter pylori and Lactococcus lactis is applied in the biological field to achieve considerable social and economic benefits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
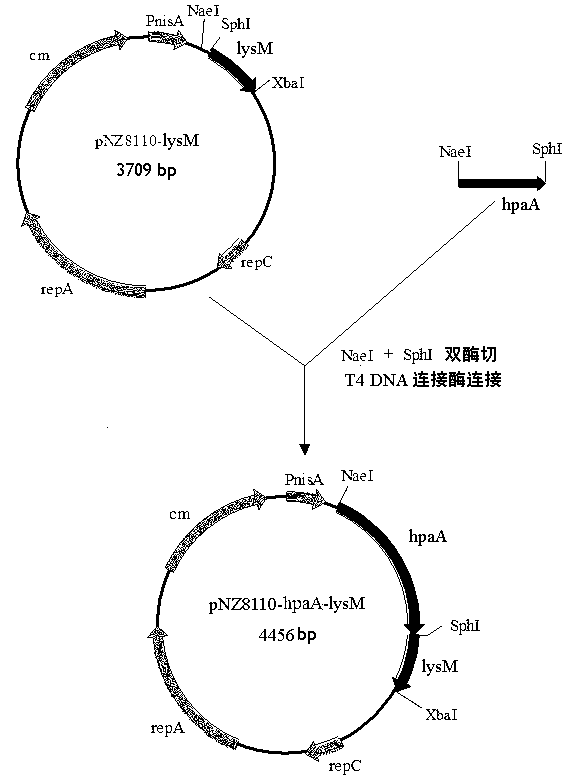
[0053] Example 1 Construction of Lactococcus lactis expression vector pNZ8110-hpaA-lysM
[0054] For the construction of Lactococcus lactis expression vector pNZ8110-hpaA-lysM see figure 1 .
[0055] 1 Preparation of Helicobacter pylori genomic DNA
[0056] 1.1 Use Brooke's blood plate medium, at 37 ° C, 5% CO 2 , under microaerophilic conditions, culture the Helicobacter pylori MEL-HP27 strain for 3-4 days, scrape 1-2 rings of bacteria with an inoculation loop, add 0.5ml sterilized deionized water and 100μl 100g / L SDS solution, boil at 100℃ for 5min .
[0057] 1.2 Add RNase A (to a final concentration of 50 μg / ml) in a 37°C water bath for 1 hour, then add proteinase K to a final concentration of 50 μg / ml, and place in a 42°C water bath for 1 hour.
[0058] 1.3 Use equal volumes of phenol, phenol-chloroform (a mixture of equal volumes of phenol and chloroform), and chloroform to extract once respectively.
[0059] 1.4 Add 2 times the volume of ice-cooled absolute ethanol...
Embodiment 2
[0109] Example 2 Construction of L.lactis NZ3900 / pNZ8110-hpaA-lysM
[0110] 1. Preparation of Competent Cells of Lactococcus lactis L.lactis NZ3900 Strain
[0111] (1) Inoculate 5ml GSGM with 200μl bacterial solution from the L.lactis NZ3900 strain tube stored at -80℃ 17 culture medium, put CO 2 Incubator, 30°C, 5% CO 2 Stand overnight culture;
[0112] (2) Add 5ml of the overnight cultured bacterial solution to 50ml of GSGM 17 medium, 30°C, 5% CO 2 Static culture for 12h~14h;
[0113] (3) Add 50ml cultured bacteria solution to 400ml GSGM 17 Medium, 30°C, 5% CO 2 static culture to OD 600 about 0.3;
[0114] (4) Centrifuge the bacterial liquid at 4°C and 4000r / min for 20min to collect the bacterial cells;
[0115] (5) Wash once with ice-cold 400ml lotion I (0.5mol / L sucrose, 100ml / L glycerol), centrifuge at 4000r / min, 4°C for 20min, and collect the bacterial solution;
[0116] (6) Resuspend the bacteria in 200ml ice-cold washing solution II (0.5mol / L sucrose, 100ml / L...
Embodiment 3
[0133] Example 3 Preparation of recombinant strain L.lactis NZ3900 / pNZ8110
[0134] The preparation of the recombinant strain L.lactis NZ3900 / pNZ8110 can refer to the technical method provided by the seller of the L.lactis NZ3900 strain, NIZO Food Research in the Netherlands or Mobitec in Germany, or the following method:
[0135] 1. Preparation of L.lactis NZ3900 Competent Cells
[0136] Method is with embodiment 2.
[0137] 2. Electrotransformation of L.lactis NZ3900 strain competent cells
[0138] Take 1 μl of the expression vector pNZ8110 (10ng~20ng) and mix it with 40 μl of L.lactis NZ3900 competent cells, and refer to Example 2 for the electrotransformation of L.lactis and the screening method for positively transformed bacteria.
[0139] 3. Identification of L. lactis positive transformants
[0140] 3.1 GM containing 10 μg / ml chloramphenicol from screening positive transformants 17 Pick a single colony on the culture plate and inoculate it in 5ml liquid GM 17 Mediu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com