Vitis bellula anthocyanidin reductase gene, protein coded by same and application of vitis bellula anthocyanidin reductase gene
A grape flower and reductase technology is applied in the fields of molecular biology, plant genetic engineering, and biology, and achieves good application prospects, improved crop resistance to diseases and insect pests, and improved forage varieties.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Embodiment 1, the synthesis of beautiful grape cDNA in South China
[0041] 1. Extraction and detection of total RNA from Meili grape in South China
[0042] Take 2 g of young leaves of Vitis bellula in South China, quickly grind them into powder with liquid nitrogen in a mortar, and quickly transfer them to 10 mL of extraction buffer preheated at 65 °C [CTAB (W / V) 2%, Tris-HCl (pH8.0) 100mmol / L, EDTA25mmol / L, NaCl2.0mol / L, PVP402%, spermidine 0.5g / L, mercaptoethanol 2%], shake and mix well; extract twice with equal volume of chloroform, Centrifuge at 7500g for 15min. Add 1 / 4 volume of 10M LiCl to the supernatant, mix and place at 4°C to precipitate overnight; centrifuge at 7500g for 20min, and use 500μL SSTE [SDS0.5%, NaCl1mol / L, Tris-HCl (pH8.0) 10mmol / L, EDTA1mmol / L], dissolved at 65°C for 5min. Extract with an equal volume of chloroform, centrifuge at 13,000g for 5min, add 2 times the volume of absolute ethanol to the supernatant, place at -70°C for 2h, centrifuge ...
Embodiment 2
[0045] Embodiment 2, the cloning of anthocyanin reductase gene
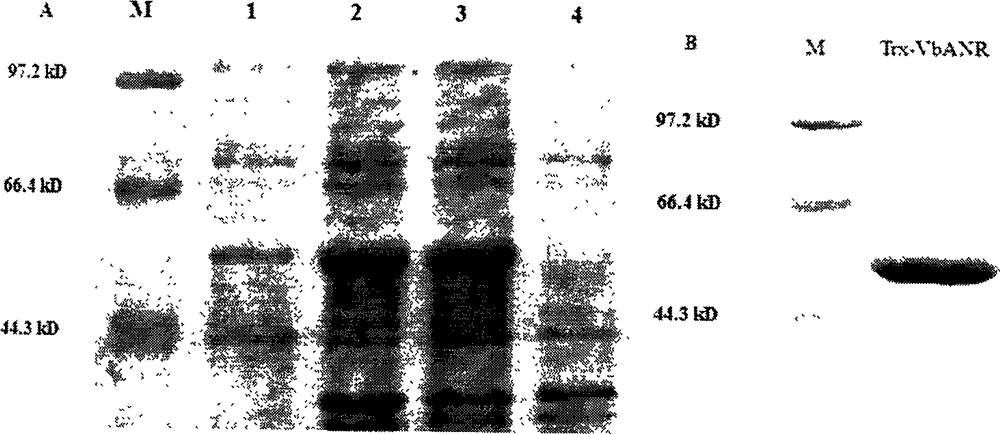
[0046] The nucleotide sequence and EST sequence related to the anthocyanin reductase gene were searched from NCBI, and a pair of amplification primers ANR-F (ATGGCCACCCAGCAC CCCAT) and ANR-R (TCAATTCTGCAATAGCCCCTTGGC) were designed using vector NTI software through comparison analysis. Using the cDNA of Huanan Meili grape as a template, follow the following system [10×Ex PCR buffer 5.0μL, dNTP (2.5mM) 4.0μL, primers LDC-F and LDC-R (10pmol) each 1.0μL, Ex Taq polymerase 0.25μL , cDNA4.0μL, add ddH 2 0 to a final volume of 50.0 μL] to amplify the LDC gene; reaction program: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 52°C for 30 s, extension at 72°C for 1 min, 30 cycles; final extension at 72°C for 10 min, and storage at 4°C. The amplification results were detected by 1% agarose gel electrophoresis ( figure 1 ). Utilize the Agarose Gel DNA Fragment Recovery Kit Ver.2.0 of TaK...
Embodiment 3
[0047] Embodiment 3, prokaryotic expression of anthocyanin reductase gene
[0048] 1. Construction of prokaryotic expression vector
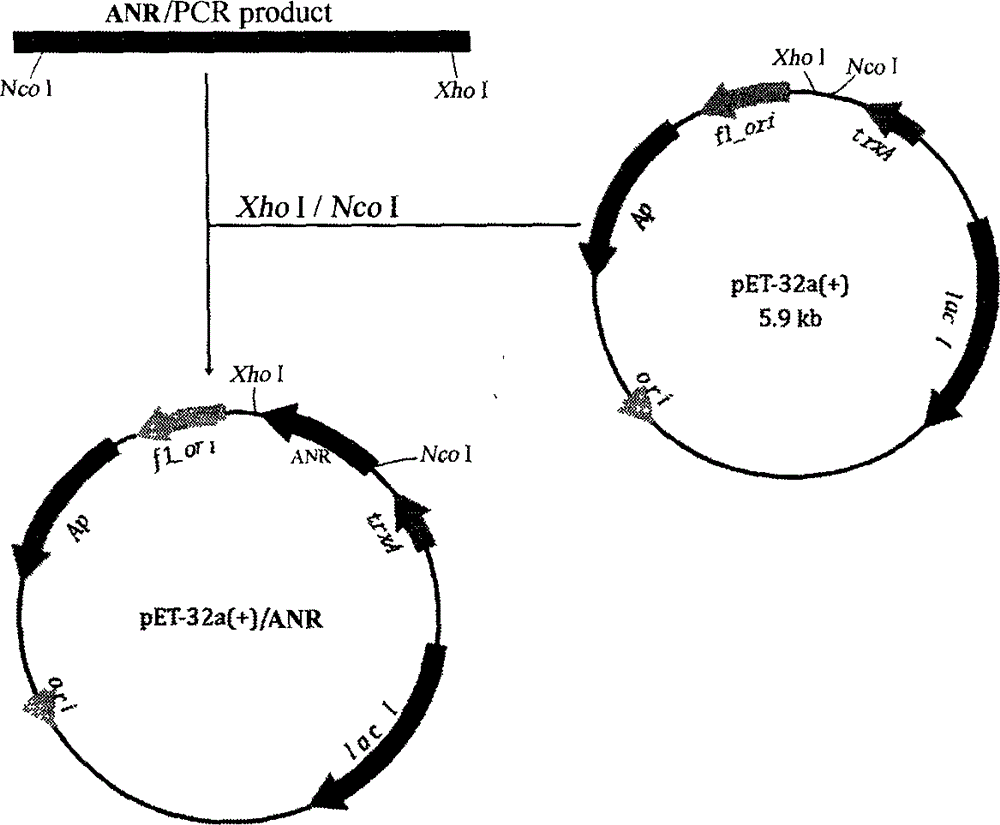
[0049] Using the ANR in the T vector as a template, primer P1 (CATG CCATGG CCACCCAG CACCCCAT) and P2 (CCG CTCGAG TCAATTCTGCAATAGCCCCTTG) for PCR reaction, the amplified product was detected and purified by electrophoresis, digested with restriction endonucleases Nco I and Xho I at 37°C for 2 h, and the digested product was purified using a recovery kit (Takara Company, China); at the same time Use Nco I and Xho I to digest the pET-32a(+) vector at 37°C for 2 hours, and use the kit to recover the large fragment.
[0050] Mix the recovered two fragments, react at 16°C for 12 hours under the action of DNA ligase, and use CaCl to ligate the product 2 Transform Escherichia coli BL21(DE3) by using the method, carry out resistance screening on ampicillin and chloramphenicol resistance plates, and conduct whole-bacteria PCR detection on the clones,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com