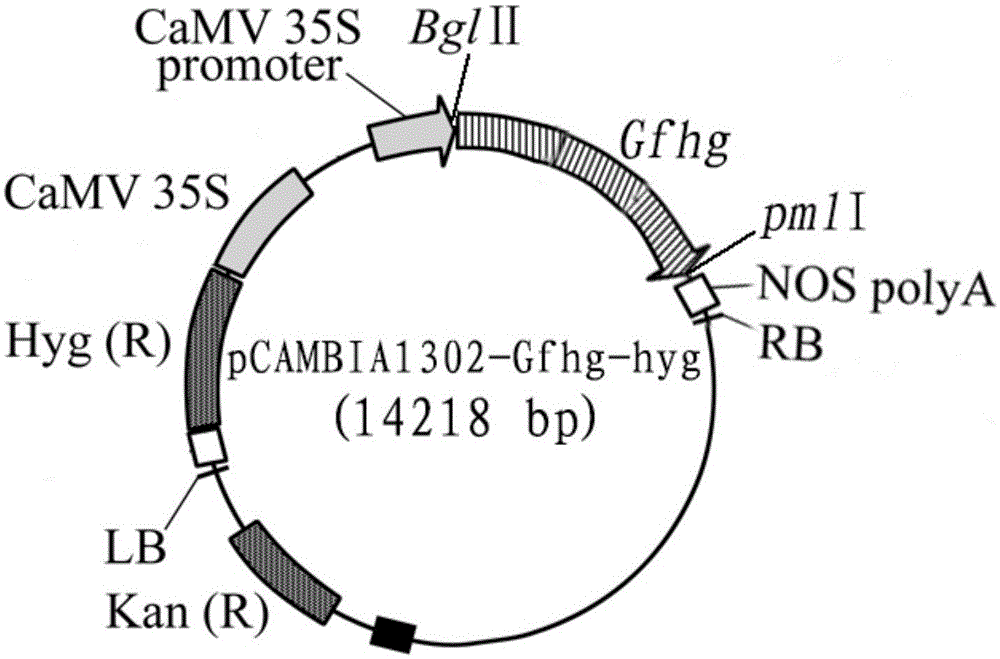
3-hydroxy-3-methylglutaryl-CoA reductase and coding gene and application of 3-hydroxy-3-methylglutaryl-CoA reductase
A methylglutaryl coenzyme and reductase technology, applied in the field of genetic engineering, can solve problems such as no cloning, and achieve the effect of improving disease resistance and stress resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] The extraction of embodiment 1 Grifola frondosa total RNA
[0029] The extraction steps are as follows:
[0030] (1) Cracking of the sample: take 50-100 mg of Grifola frondosa fruiting body and put it in a mortar, add liquid nitrogen to quickly grind it into powder, transfer it into a centrifuge tube that has added 1 ml Trizol, and blow and mix with a pipette;
[0031] (2) Stand at room temperature for 5 minutes, centrifuge at 12,000 rpm for 10 minutes at 4°C;
[0032] (3) Transfer the supernatant to another centrifuge tube, add 200 μL of chloroform, mix by inversion, place at room temperature for 10 min, and centrifuge at 12000 rpm for 15 min at 4 °C;
[0033] (4) Transfer the upper aqueous phase, move it to another centrifuge tube, add an equal volume of isopropanol, mix well, and place at room temperature for 5-10 minutes;
[0034] (5) Centrifuge at 12000rpm for 10min at 4°C, discard the supernatant;
[0035] (6) Add 500 μL of 75% ethanol, mix for a while, centrif...
Embodiment 2
[0037] Cloning of the 3-hydroxyl-3-methylglutaryl-CoA reductase gene (Gfhg) of Example 2 Grifola frondosa
[0038] Take 1 μg of total RNA, use randomhexamers as primers, follow the instructions of SuperScriptIII First-StrandSynthesisSystem, and reverse transcribe to generate the first strand.
[0039] Primers were designed according to the CDS sequence of Grifola frondosa 3-hydroxy-3-methylglutaryl-CoA reductase gene as follows:
[0040] Upstream primer SEQ ID NO.1Gfhg-F: gaagatctatgcgtgcgatacttcgcccgc;
[0041] Downstream primer SEQ ID NO.2Gfhg-R: cccacgtgtcacttcgtctccacagcatagc;
[0042] The first-strand cDNA generated by reverse transcription of the total RNA of Grifola frondosa was used as a template for PCR amplification.
[0043] The PCR reaction system is as follows:
[0044]
[0045] The PCR amplification procedure is as follows:
[0046]
[0047] Refer to the instructions of QuickGelExtraction Kit (Kangwei Century) to perform gel extraction and recovery of t...
Embodiment 3
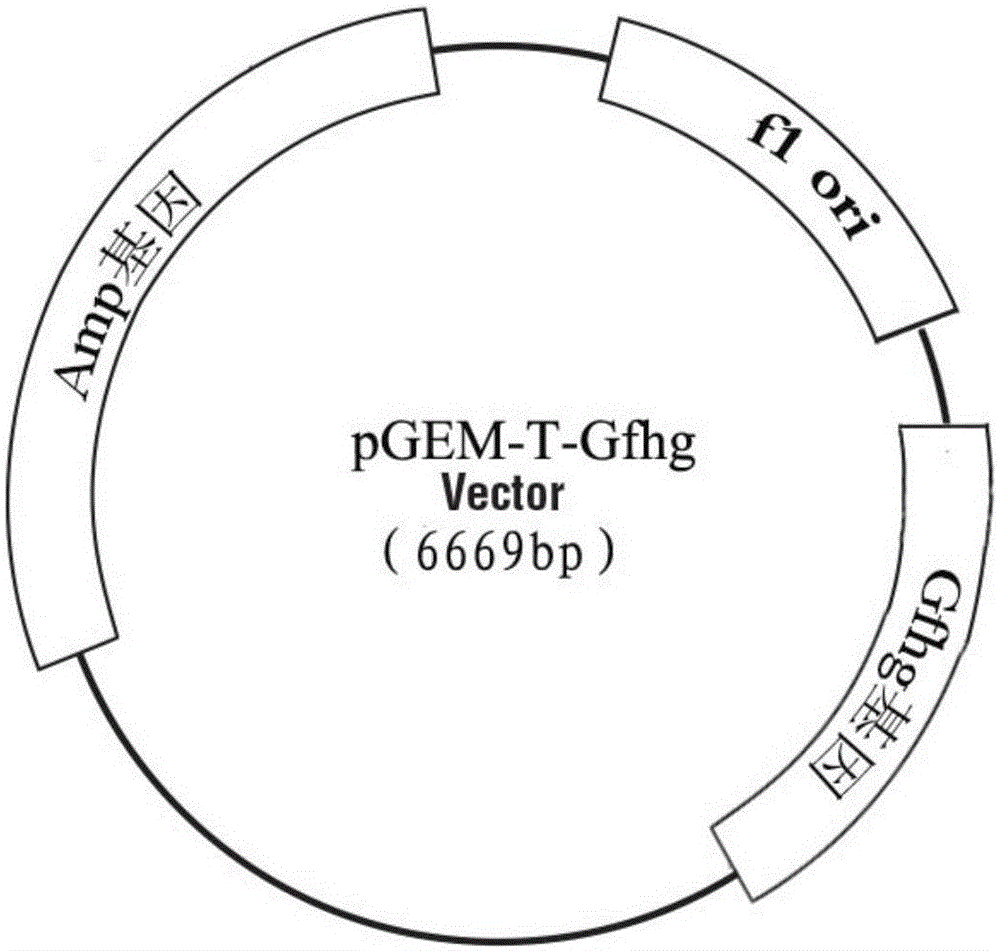
[0048] Construction of embodiment 3 cloning vector pGEM-T-Gfhg
[0049] Referring to pGEM-TVectorSystems (Promega), T4DNALigase in the kit was used to connect the Gfhg gene fragment (SEQ ID NO.5) recovered from gel cutting to the T vector.
[0050]Referring to the instructions of pGEM-TVectorSystems (Promega), the ligation product was transformed into DH5α Escherichia coli competent cells. Add 400 μL LB liquid medium to the transformation product, culture at 37°C for 2 hours on a shaker at 200 rpm, spread evenly on the LB solid culture dish containing X-Gal, IPTG, and AMP, place it upright for 1 hour to dry, and culture it upside down at 37°C for 28 -24h to form a single colony.
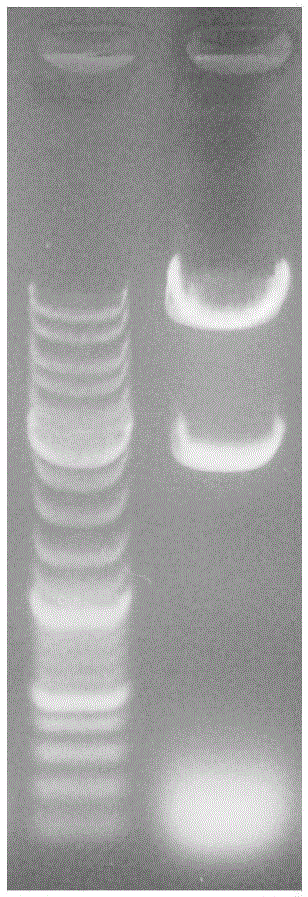
[0051] Use the above primers Gfhg-F and Gfhg-R to identify 10 single clones by PCR. PCR conditions: 93°C, 3min; 93°C, 30s; 56°C, 30s; 72°C, 3min, 30 cycles; 72°C, 10min . PCR product electrophoresis verification, select the clone with the correct electrophoresis band and send it to Shanghai Sangon...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com