Biosynthetic gene cluster of paquete amide and application thereof
A technology of pactamide and biosynthesis, applied in the direction of plant genetic improvement, application, microorganisms, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0164] Extraction of Streptomyces pactum SCSIO 02999 total DNA:
[0165] The mycelium of fresh Streptomyces pactum (Streptomyces pactum) SCSIO 02999 was inoculated into 50 mL of TSB medium (17 g of tryptone, 3 g of plant peptone, 5 g of sodium chloride, 2.5 g of dipotassium hydrogen phosphate, 2.5 g of glucose) according to an inoculum size of 5%. g, add water to 1 L, pH 7.2-7.4), 28-30 ° C, shaking culture for about 3-4 days, 4000 rpm for 10 minutes to collect mycelia. Wash the mycelia twice with STE solution (NaCl 75mM, EDTA 25mM, Tris-Cl 20mM), add 30mL of STE solution and lysozyme with a final concentration of 3mg / mL to the washed mycelium, vortex evenly, and store at 37°C Incubate for 3 hours, add proteinase K to a final concentration of 0.1-0.2mg / mL, mix well, incubate at 37°C for 10 minutes, add SDS to a final concentration of 1-2%, mix well, put in a 55°C water bath for about 1 hour, Invert several times during this period. Add an equal volume of phenol-chloroform-is...
Embodiment 2
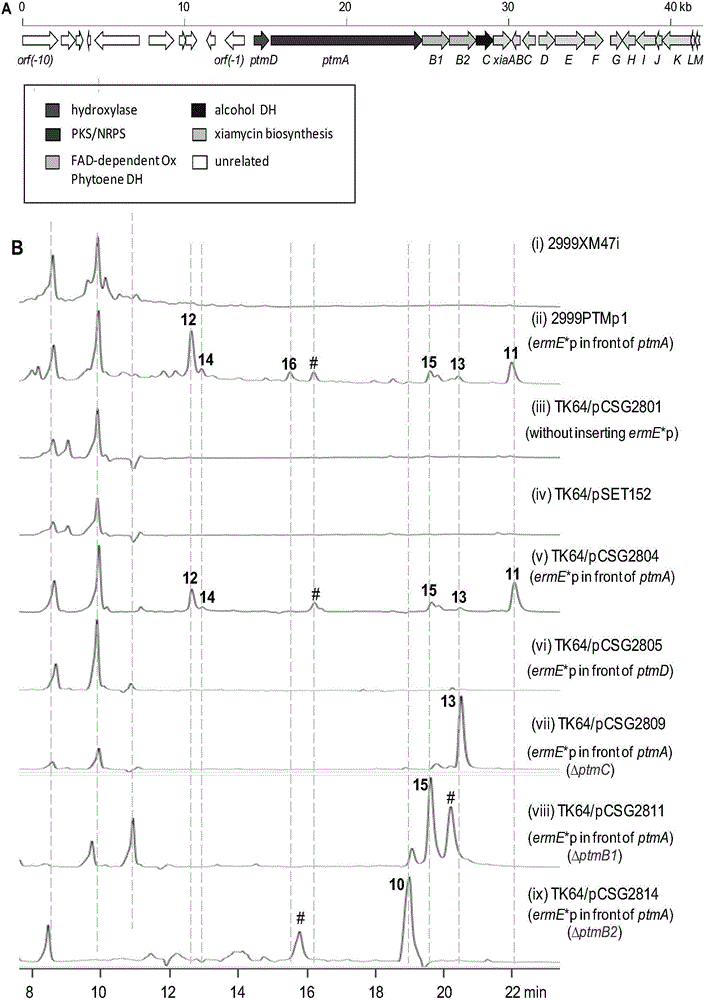
[0167] Streptomyces pactum (Streptomyces pactum) SCSIO 02999 whole genome library establishment:
[0168] First, through a series of dilution experiments to determine the amount of restriction endonuclease Sau3A I, in a 20 μL system, containing 17 μL of Streptomyces pactum (Streptomyces pactum) SCSIO 02999 total DNA, 2 μL of 10 × reaction buffer and 1 μL of different dilutions Sau3A I, the stop reaction is 4 μL 0.5mol / L EDTA and the appropriate loading buffer. Through exploration, it is determined that the enzyme activity unit of 0.025-0.05U is more appropriate. On this basis, a genomic DNA fragment slightly larger than 40kb was obtained by a large number of partial enzyme digestions, and dephosphorylated with a dephosphorylase.
[0169] The vector SuperCos l plasmid used to construct the library was first cut from the middle of the two cos sequences with restriction endonuclease Xba I, then dephosphorylated, and then used restriction endonuclease Bam from the multiple clonin...
Embodiment 3
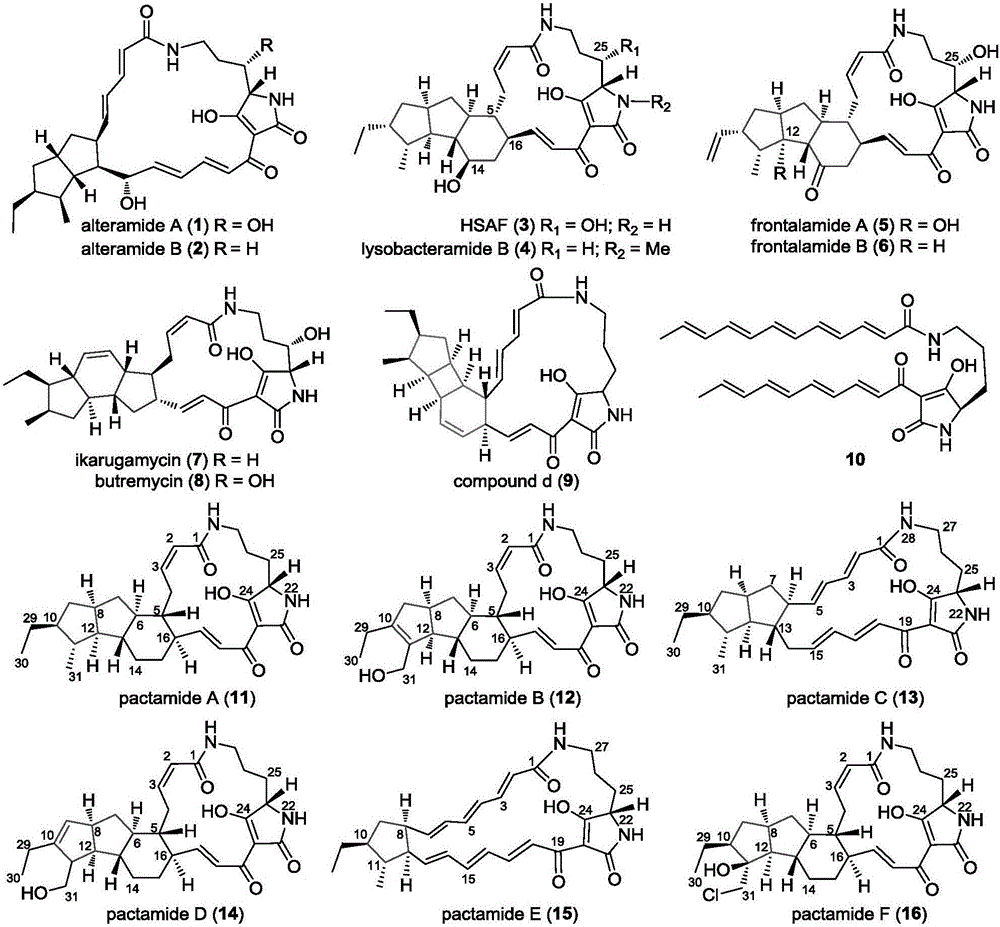
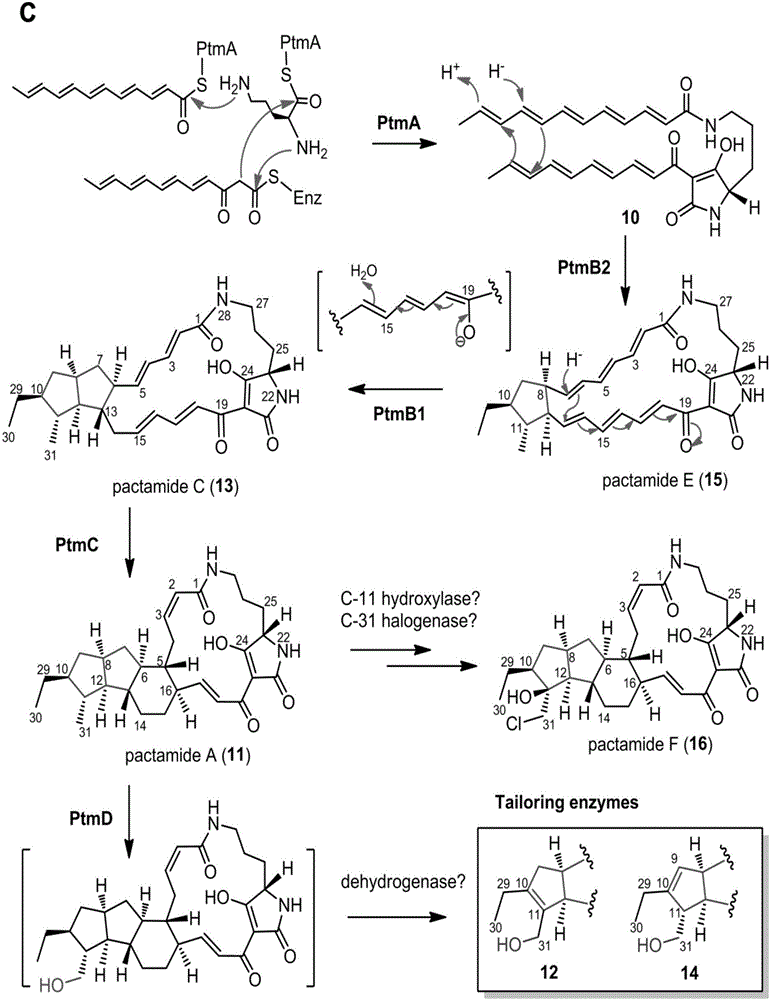
[0171] Example 3: Determination of cytotoxic activity of pactamide A-F
[0172] The activity of pactamide A-F was measured against four tumor cell lines: human breast cancer MCF-7 cells, human liver cancer cells HepG2, human lung cancer H460 cells and human glioma cells SF-268. For experimental methods, refer to [Wu, Z.C.; Li,D.L.; Chen,Y.C.; Zhang,W.M.,A new isofuranonaphthalenone and benzopyransfrom the endophytic fungus Nodulisporium sp.A4from Aquilariasinensis.Helv.Chim.Acta 2010,93,(5),920-924.] ) as a control, its IC to human cancer cells including breast cancer MCF-7, human lung cancer H460 cells, human liver cancer HepG2 and human glioma cells SF-268 50 They are 9.2 μM, 1.5 μM, 1.4 μM and 3.9 μM, respectively. The results showed that the IC of pactamide A on human cancer cells including breast cancer MCF-7, human lung cancer H460 cells, human liver cancer HepG2 and human glioma cells SF-268 50 0.26μM, 0.24μM, 0.37μM and 0.51μM respectively; the IC of pactamide B on h...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com