Recombinant saccharomycetes with soybean hull peroxidases displayed on cell surfaces as well as construction method and application of recombinant saccharomycetes
A technology of bean shell peroxide and bean shell peroxidase, which is applied in the field of genetic engineering, can solve the problems of low host cells, difficulty in purification of secreted and expressed ShP, and complicated separation and purification process of natural ShP, so as to overcome the cumbersome separation and purification operation. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Soybean shell peroxidase surface display plasmid construction, the steps are as follows:
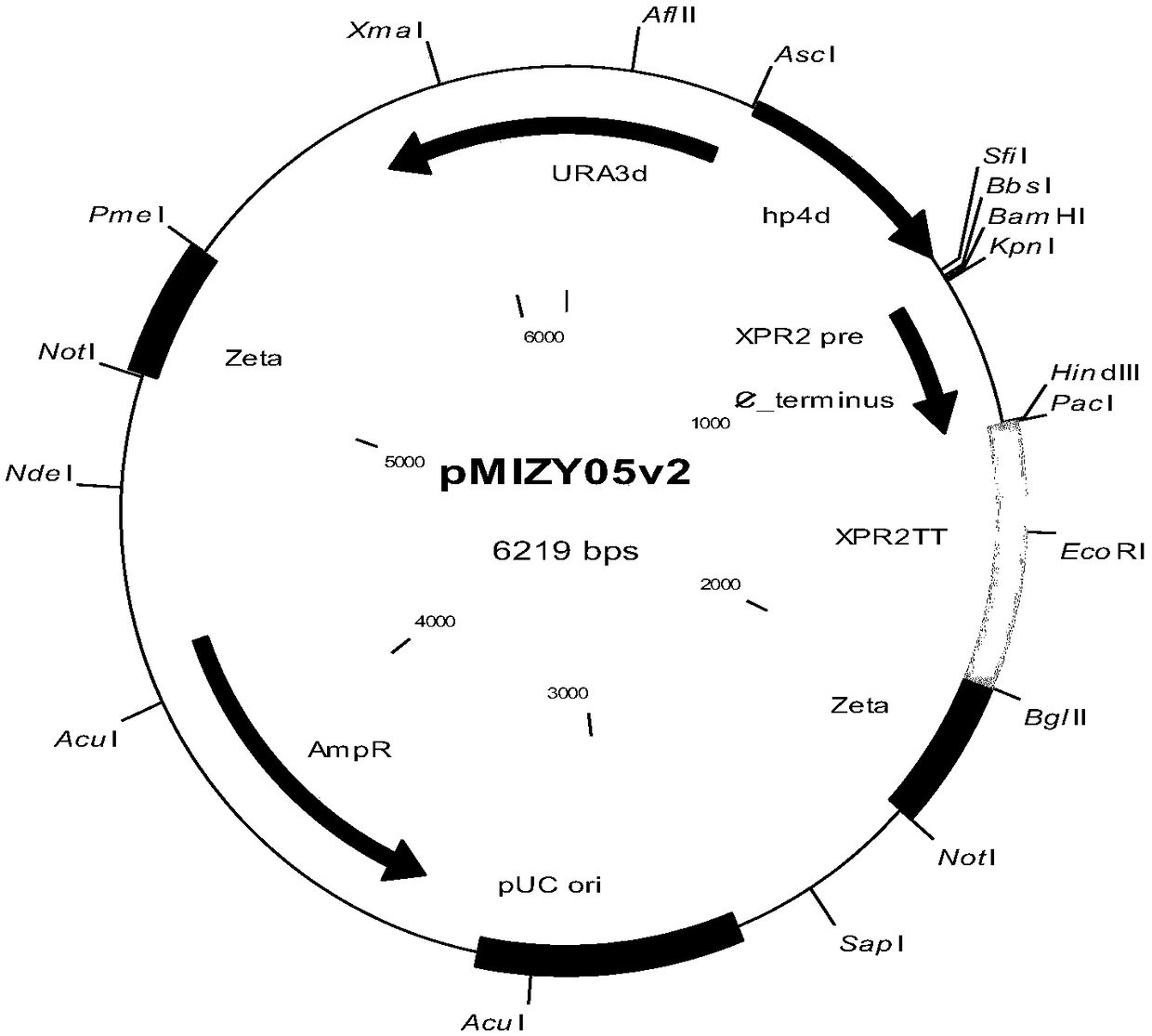
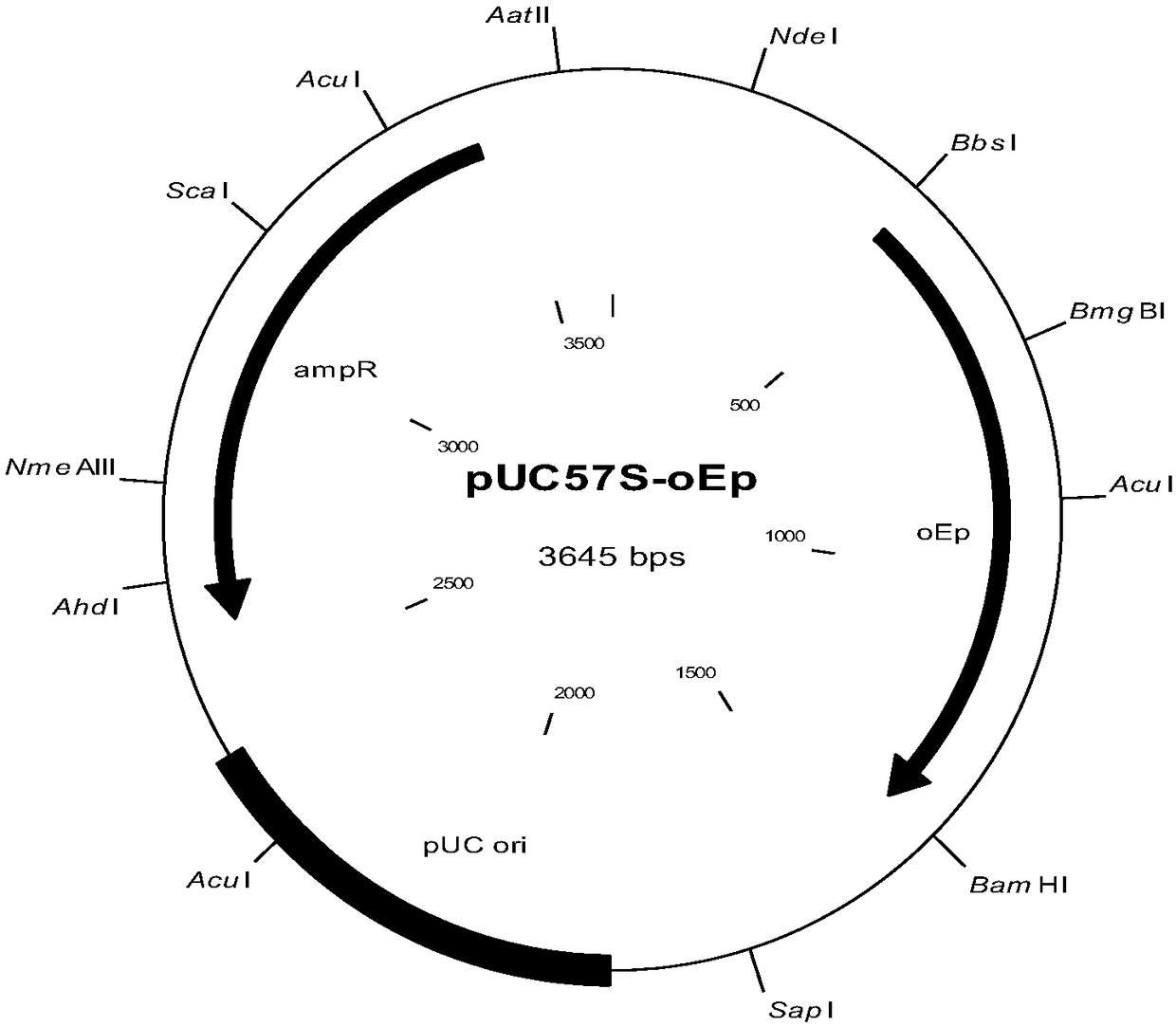
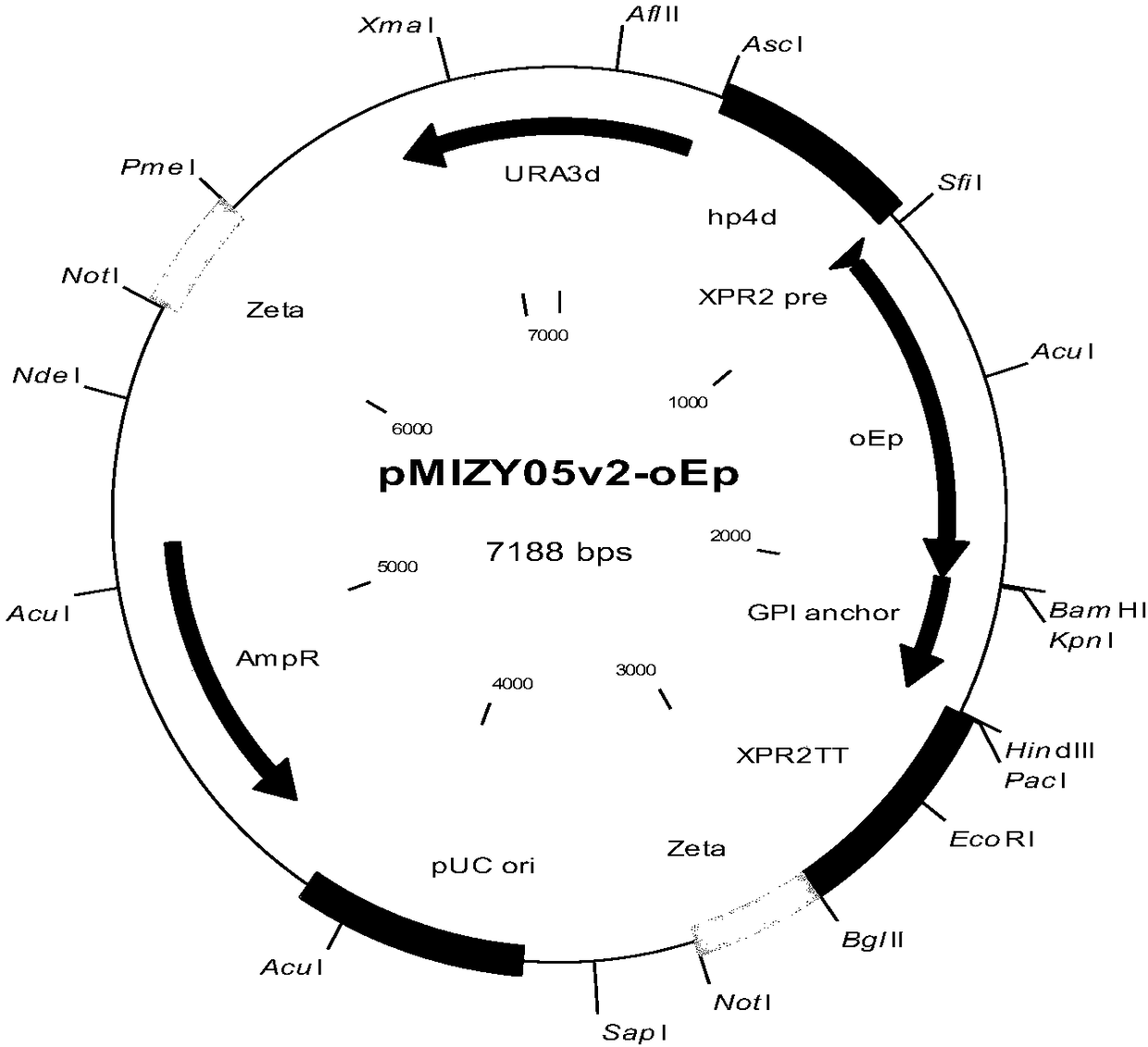
[0052] Design the nucleotide sequence shown in SEQ ID No.1, and send it to a third-party company to synthesize and clone it into the pUC57-simple vector to obtain the pUC57S-oEp plasmid (plasmid map as shown in figure 2 shown), after being digested with SfiI / BamHI restriction endonuclease, the target fragment was recovered and connected to the pMIZY05v2 expression vector digested with the same enzyme (plasmid map as shown in figure 1 shown) above.
[0053] SEQ ID NO.1 (the underlined parts are SfiI and BamHI restriction enzyme sites respectively):
[0054] GT GGCCGTTCTGGCCCAGCTGACCCCCACCTTCTACCGAGAGACCTGTCCCAACCTGTTCCCCATCGTGTTCGGCGTCATTTTCGACGCCTCTTTCACCGACCCCCGAATCGGAGCTTCCCTGATGCGACTGCACTTCCACGACTGTTTCGTGCAGGGTTGCGACGGCTCTGTCCTGCTGAACAACACCGACACCATCGAGTCTGAGCAGGACGCCCTGCCCAACATCAACTCCATTCGAGGACTGGACGTGGTCAACGACATTAAGACCGCTGTGGAGAACTCTTGTCCCGACACCGTCTCCTGCGCCGACATCCTGGCTATTG...
Embodiment 2
[0073] Obtaining of recombinant strains, the steps are as follows:
[0074] (1) Preparation of recombinant fragments The pMIZY05v2-oEp recombinant plasmid was digested with NotI restriction endonuclease and detected by agarose gel electrophoresis. After digestion of pMIZY05v2-oEp, two fragments are expected to appear, the sizes are 4.5kbp and 2.7kbp respectively. Recombinant vector NotI digestion and electrophoresis results are as follows: image 3 As shown, the two fragments are consistent with expectations. Recycle large fragments, spare
[0075] (2) Yeast Competent Cell Preparation Inoculate the Yarrowia lipolytica PO1h strain on YPD solid medium and culture overnight at 28°C; pick a single colony and inoculate it in 5.0mL liquid YPD medium, and culture overnight at 28°C with shaking ; at a final concentration of 5.0×10 6 cells / mL were inoculated in 50.0 mL liquid YPD medium, cultured with shaking at 200 rpm for 4 h at 28 °C; centrifuged at 1500 × g for 5 min at 4 °C to...
Embodiment 3
[0079] To verify the ability of recombinant strains to produce immobilized enzymes, the steps are as follows:
[0080] (1) Pick a single colony of recombinant yeast and inoculate it in 5.0 mL of YPD liquid medium, culture it with shaking at 28°C overnight, and this is the seed solution.
[0081] (2) Take 1.0 mL of seed liquid, transfer it to 50.0 mL of PPB medium, culture it with shaking at 25°C for 3-5 days, and the obtained fermentation liquid is the crude enzyme liquid.
[0082] (3) The activity of ShP in the crude enzyme solution was detected by ABTS method, the darker the color, the higher the activity, and thus the strains with high activity were screened. For the darker color, determine the specific value of ShP activity, and the reaction solution system is as follows:
[0083]
[0084] Incubate at 35°C and pH 6.0 for 5 minutes, and the color changes after the reaction is as follows: Figure 4 shown (green). Measure the change of the 420nm absorbance value of the ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com