Recombinant fibronectin with anti-wrinkle repairing function and preparation method and application of recombinant fibronectin
A fibronectin and functional technology, applied in the field of its preparation and recombinant fibronectin, can solve the problems of difficulty in removing His-tag, no recombinant fibronectin, insufficient specificity, etc., and achieve a wide salt ion concentration Tolerance range, excellent temperature stability range, high purity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1: Recombinant fibronectin with anti-wrinkle repair function
[0051] The invention relates to a recombinant fibronectin with anti-wrinkle repair function, the recombinant fibronectin sequence includes at least one RGD domain that can bind to cell integrin receptors, at least one section that can act on sodium ion channels and bind to sodium ion channels A peptide segment, the RGD structural domain and the sodium ion channel binding peptide segment are connected by a polypeptide linking segment comprising 0-300 amino acid residues.
[0052] Based on the function of natural conus peptide, its Mu-CnIIIC fragment is selected as the sodium ion channel binding peptide, and the amino acid sequence of the Mu-CnIIIC fragment is QGCC XXXXX css XX C B DH Z RCCGRR, X in the amino acid sequence of the Mu-CnIIIC fragment is any one of 20 amino acid residues, namely glycine, alanine, valine, leucine, isoleucine, phenylalanine , Proline, Tryptophan, Tyrosine, Serine, Cyst...
Embodiment 2
[0056] Example 2: Preparation method of recombinant fibronectin with anti-wrinkle repair function
[0057] 1. Preparation method of recombinant fibronectin with anti-wrinkle repair function
[0058] Include the following steps:
[0059] (1) Vector construction
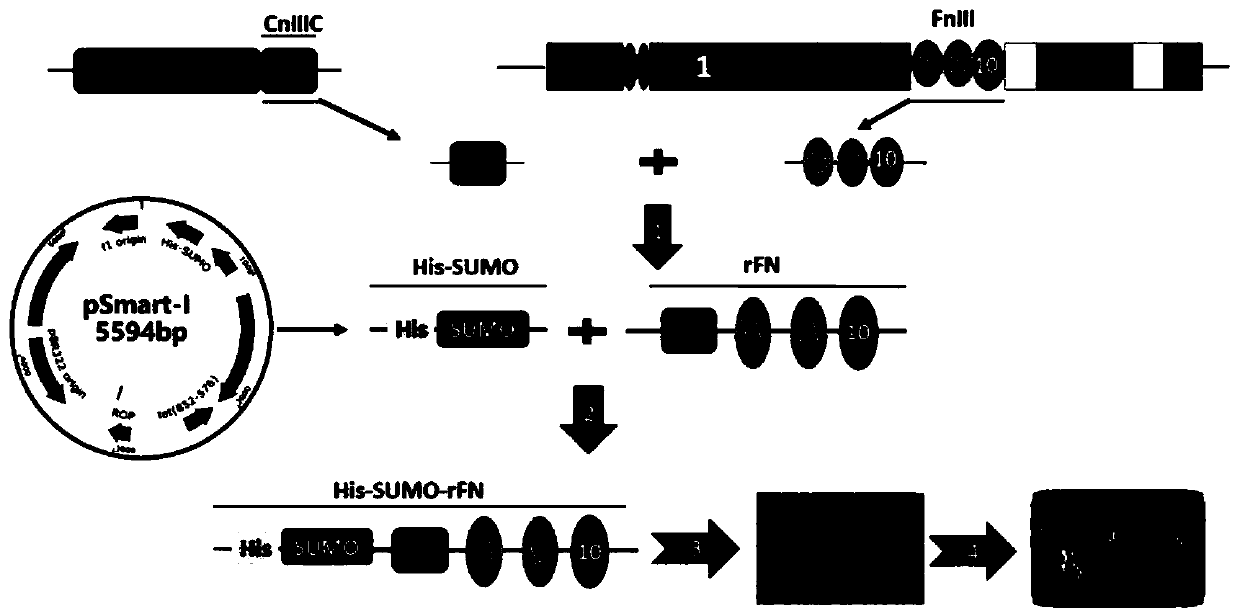
[0060] like Figure 2A Indicated by the arrow 1, the FnIII-8, FnIII-9, FnIII-10 domains of natural fibronectin are intercepted, and the Mu-CnIIIC fragment of the natural conus peptide is intercepted, and the FnIII The -10 domain is connected to the Mu-CnIIIC fragment, and the FnIII-8, FnIII-9, FnIII-10 domain and Mu-CnIIIC fragment are seamlessly spliced by whole gene optimization and synthesis to obtain the coding DNA of recombinant fibronectin rFN.
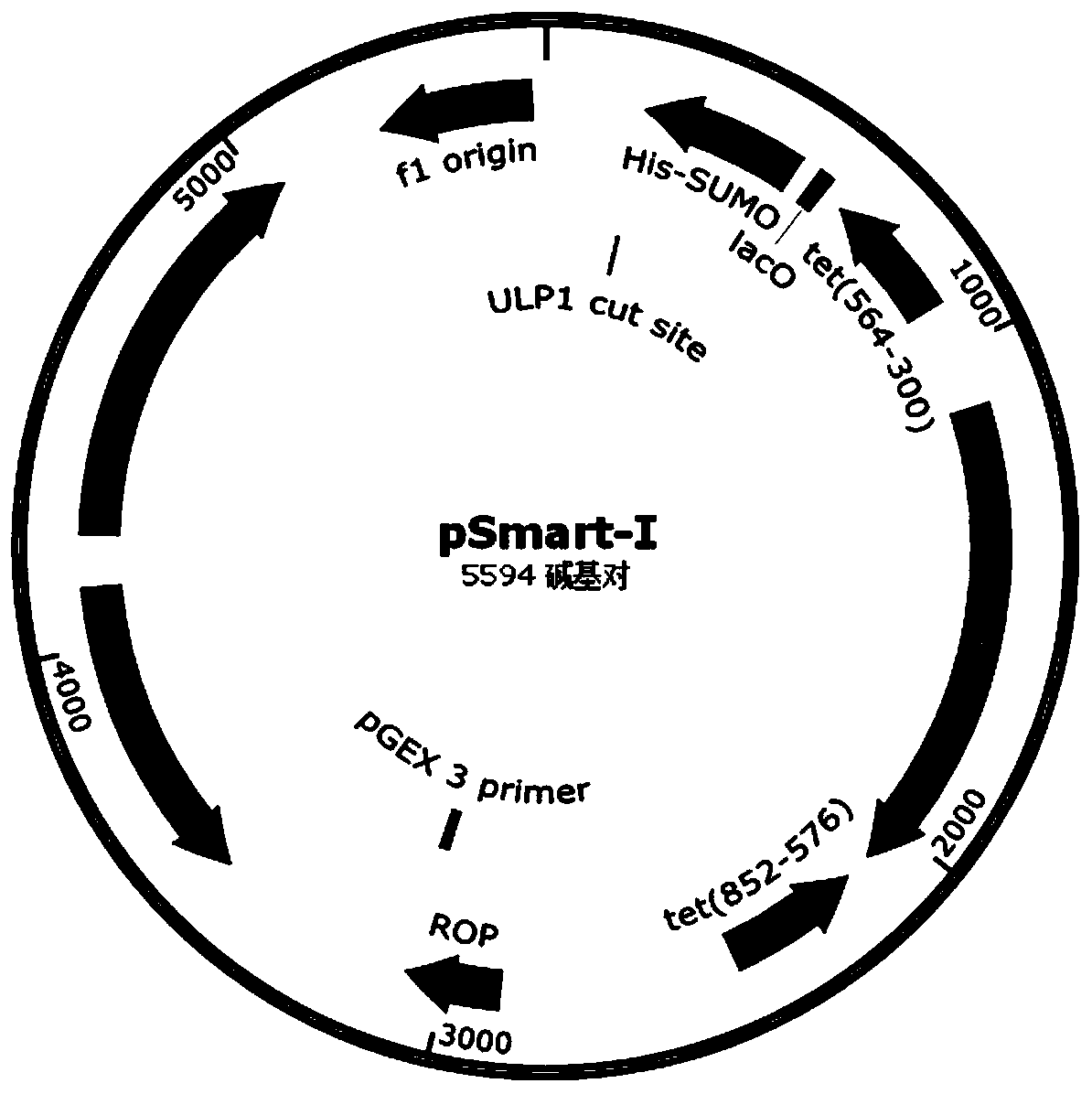
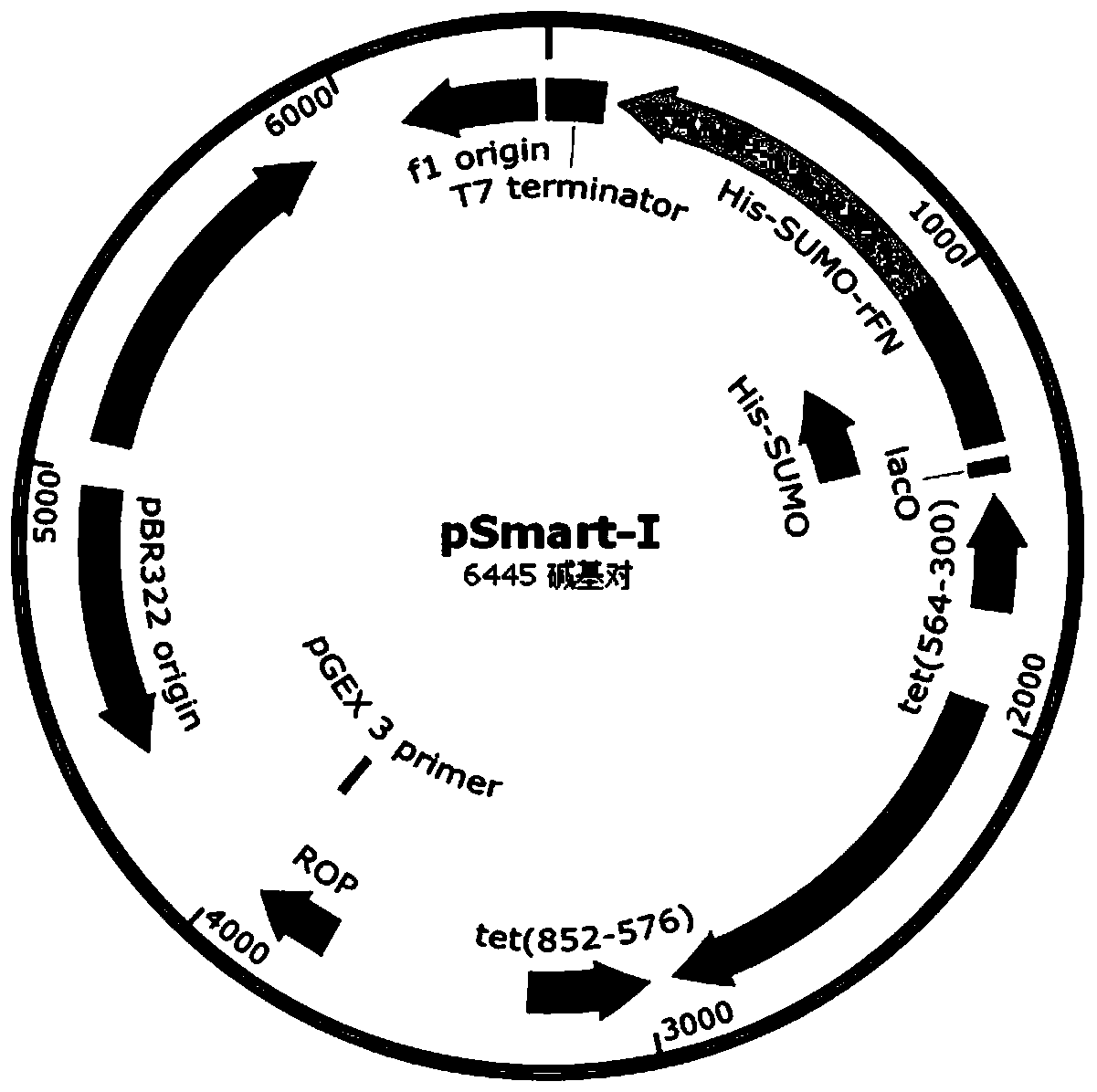
[0061] like Figure 2A Indicated by the arrow 2, the DNA encoding the recombinant fibronectin rFN was inserted into the appropriate reading frame position of the pSmart-I vector. The pSmart-I vector was purchased from Changzhou Tiandi Renhe Biotechnology Co., Ltd. ...
Embodiment 3
[0108] Example 3: Stability test of recombinant fibronectin with anti-wrinkle repair function
[0109] 1. Temperature stability test
[0110] Take 1ml of the purified recombinant fibronectin rFN sample and divide it into 6 EP tubes, place them in water baths at 25°C, 40°C, 50°C, 60°C and 70°C for incubation for 3 hours, centrifuge at 12000rpm for 5min after the water bath is over Afterwards, the supernatants were taken for SDS-PAGE electrophoresis detection.
[0111] The result is as Figure 7A As shown, the first well was incubated at 25°C, and the second, third, fourth, and fifth wells were incubated at 40°C, 50°C, 60°C, and 70°C respectively. M is the pre-stained molecular weight protein marker. The results show: The heat resistance of zonulin rFN is good, no centrifugation precipitation was observed at 25°C for 3 hours, and a small amount of centrifugation precipitation was produced at 40°C, 50°C, 60°C, 70°C for 3 hours, but the remaining recombinant fibronectin in the sup...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com