Capture probe set, method and kit for detecting pathogenic microorganisms and application
A technology of pathogenic microorganisms and capture probes, applied in biochemical equipment and methods, microbial measurement/testing, combinatorial chemistry, etc., can solve the problems that mNGS hybrid capture detection products have not been established, and the effect is not significant
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] A kit for detecting pathogenic microorganisms, used for rapid hybrid capture of mNGS, designed by the following method.
[0070] 1. Design and synthesis of capture probe sets.
[0071]Review the previous clinical samples of our company, screen the pathogenic microorganisms with high clinical frequency, covering bacteria, fungi, DNA viruses, RNA viruses, parasites, and common drug resistance genes, select the coding region of pathogenic microorganisms as the target region, and based on known For the genome sequence, analyze and screen the specific regions of each species, and design and generate probe groups in the form of tile arrays. The length of the probes is about 120bp, and the probes with biotin-labeled nucleotides are synthesized.
[0072] The design of the probe requires high specificity and is not affected by homologous sequences of other closely related species. And their sequences are not affected by each other. These sequences are located in the coding reg...
Embodiment 2
[0107] A kind of pathogenic microorganism detection method, adopts the test kit described in embodiment 1, comprises the following steps:
[0108] 1. Pre-capture library preparation.
[0109] The nucleic acid of the sample to be tested is extracted, and the library is constructed so that the final library fragment is 300-600 bp, and the concentration of the library is quantitatively detected after purification.
[0110] 2. Capture by liquid phase hybridization.
[0111] 2.1 Preparation of library mixture:
[0112] Take 8 to 16 adapter libraries, and quantitatively take 125ng of each library. After mixing, add concentrated reagents according to the table below, and add the corresponding adapter blocking solution according to the library construction process of Illumina or BGI, and select according to the subsequent sequencing platform.
[0113] Table 7. Library Mixture Composition
[0114] reaction system Amount (μL) / library library to be hybridized x J...
Embodiment 3
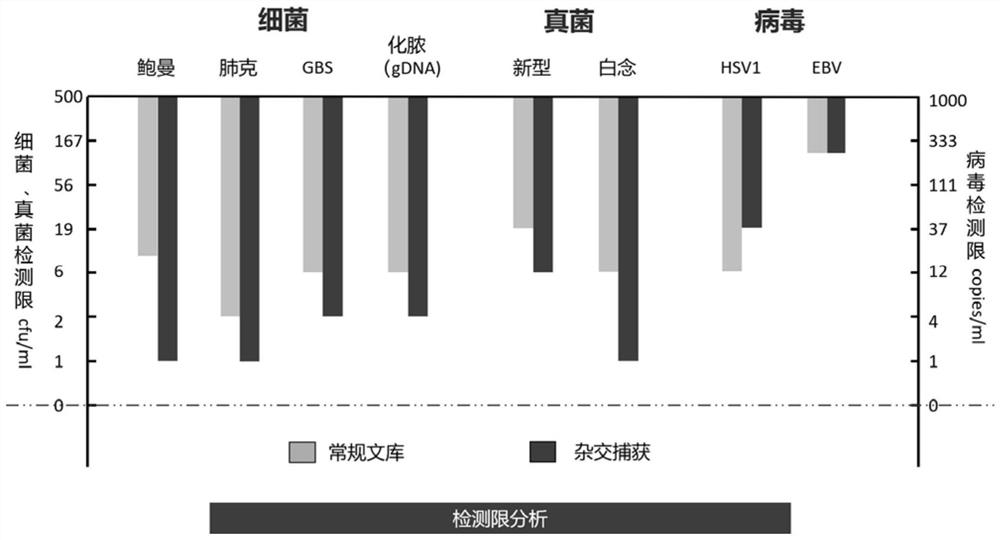
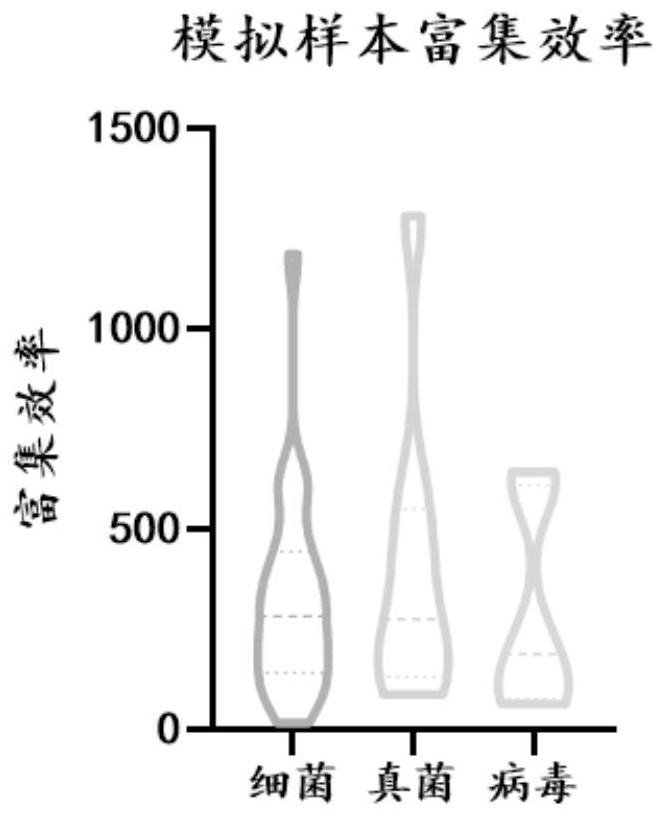
[0175] A comparative experiment of the detection limits of conventional metagenomic detection and the probe hybridization capture method of the present invention.
[0176] 1. Method.
[0177] 1.1 Prepare simulated mixed samples.
[0178] The simulated mixed sample contains human HELA cells, Acinetobacter baumannii (Baumann), Klebsiella pneumoniae (Klebsiella pneumoniae, lung grams), Streptococcus agalactiae (Streptococcus agalactiae, GBS) DNA, pyogenic Streptococcus pyogenes, pyogenes, Cryptococcus neoformans, Candida albicans, human herpesvirus type Ⅰ (HSV1), human herpesvirus type 4 (EBV).
[0179] Each mock-mixed sample contained 2 × 10 HELA cells 5 cells / ml, the prepared pathogenic microorganism mixture was diluted 3× with sterile PBS, and put into HELA cells in equal amounts, and finally the concentration of pathogenic microorganisms was obtained as shown in the table below.
[0180] Table 10. Pathogenic microorganisms and concentrations in simulated mixed samples
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com