Methods for detection or measurement of viruses
a technology for applied in the field of methods of detecting or measuring viruses, can solve the problems of not always highly sensitive or specific methods, require long procedures, and still present risk of secondary infection, and achieve the effect of simple and sensitive detection and quantitation of virus antigens
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Expression and Purification of a HCV-Derived Polypeptide
(A) Construction of an Expression Plasmid
[0122]A plasmid corresponding to the core region of HCV was constructed as follows: one microgram each of DNA of plasmids pUC·C11-C21 and pUC·C10-E12 obtained by integrating the C11-C21 clone and the C10-E12 clone (Japanese Unexamined Patent Publication (Kokai) No. 6 (1994)-38765) respectively, into pUC119 was digested in 20 μl of a restriction enzyme reaction solution [50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 100 mM NaCl, 15 units of EcoRI and 15 units of ClaI enzyme] and the restriction enzyme reaction solution [10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 50 mM NaCl, 15 units of ClaI and 15 units of KpnI enzyme] at 37° C. for one hour each, and then was subjected to 0.8% agarose gel electrophoresis to purify about 380 by of EcoRI-ClaI fragment and about 920 by of ClaI-KpnI fragment.
[0123]To the two DNA fragments and a vector obtained by digesting pUC119 ...
example 2
Method of Constructing a Hybridoma
[0132]The fusion polypeptide (TrpCll) prepared by the method described above was dissolved in 6 M urea, and then diluted in 10 mM phosphate buffer, pH 7.3, containing 0.15 M NaCl to a final concentration of 0.2 to 1.0 mg / ml, and mixed with an equal amount of adjuvant (Titermax) to make a TrpCll suspension. This suspension prepared at 0.1 to 0.5 mg / mi of TrpCll was intraperitoneally given to 4 to 6 week old BALB / c mice. Similar immunization was conducted every two weeks and after about two more weeks 10 μg of TrpCll dissolved in physiological saline was administered through the tail vein.
[0133]Three days after the last booster, the spleen was aseptically isolated from the immunized animal and was cut into pieces using scissors, which were then crumbed into individual cells and washed three times with the RPMI-1640 medium. After washing, a mouse myeloma cell line SP2 / 0Ag14 at the logarithmic growth phase as described above, 2.56×107 of said cells and ...
example 3
Construction of Monoclonal Antibody
[0136]The hybridomas obtained in the method of Example 2 were inoculated to the abdominal cavity of mice treated with pristane etc., and the monoclonal antibodies produced in the ascites fluid was collected. The monoclonal antibodies were purified using the Protein A-bound Sepharose column to separate IgG fractions.
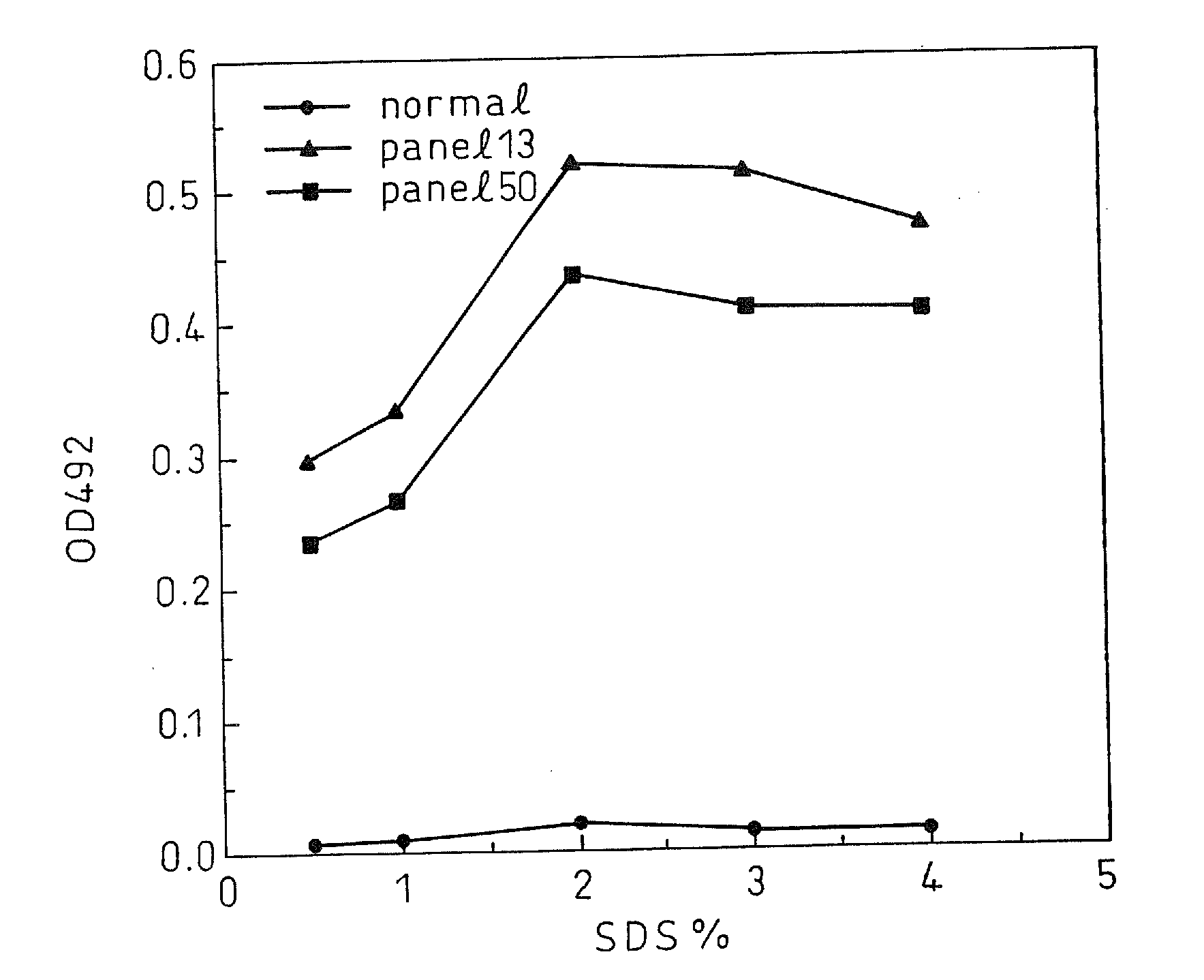
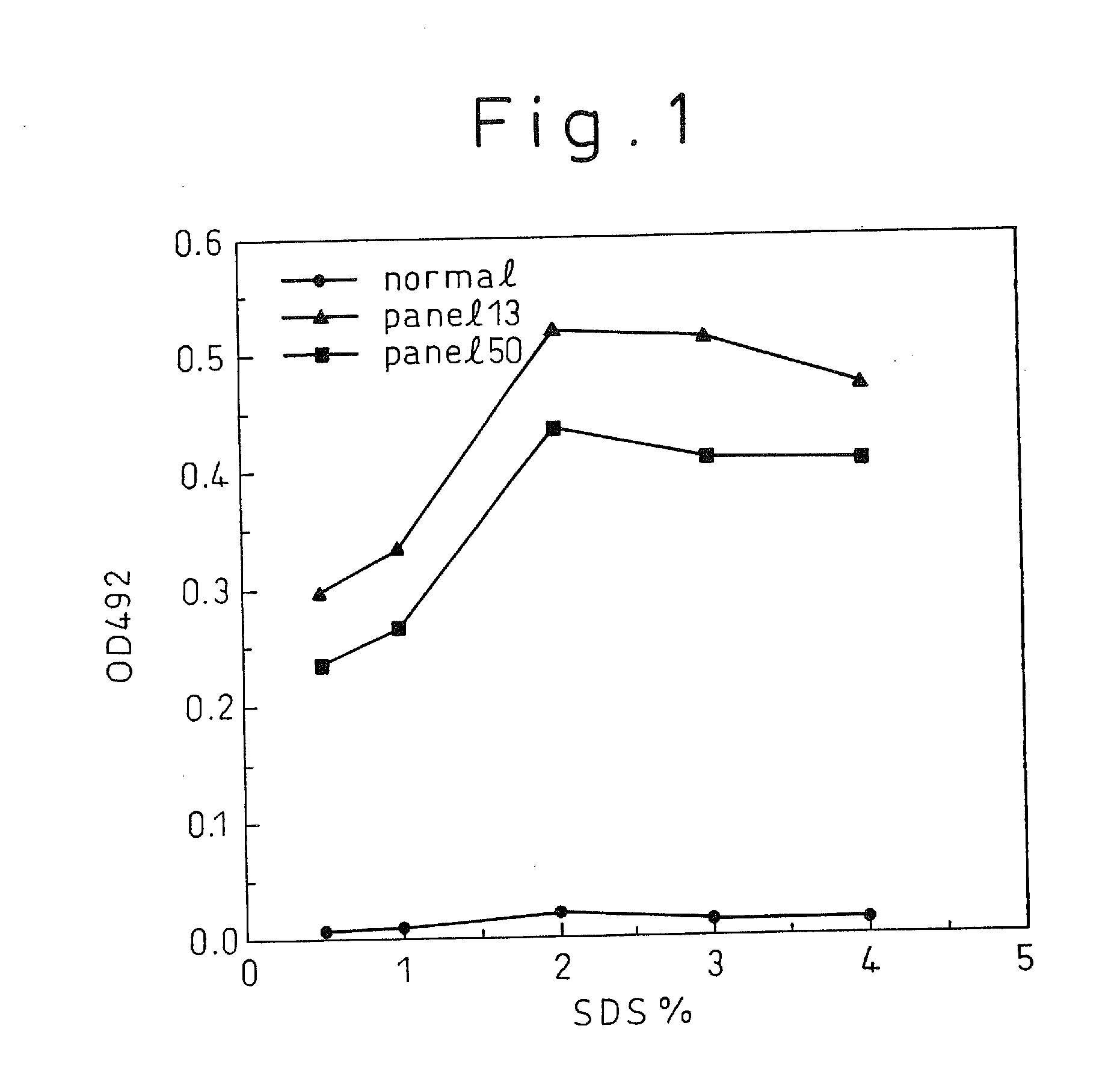
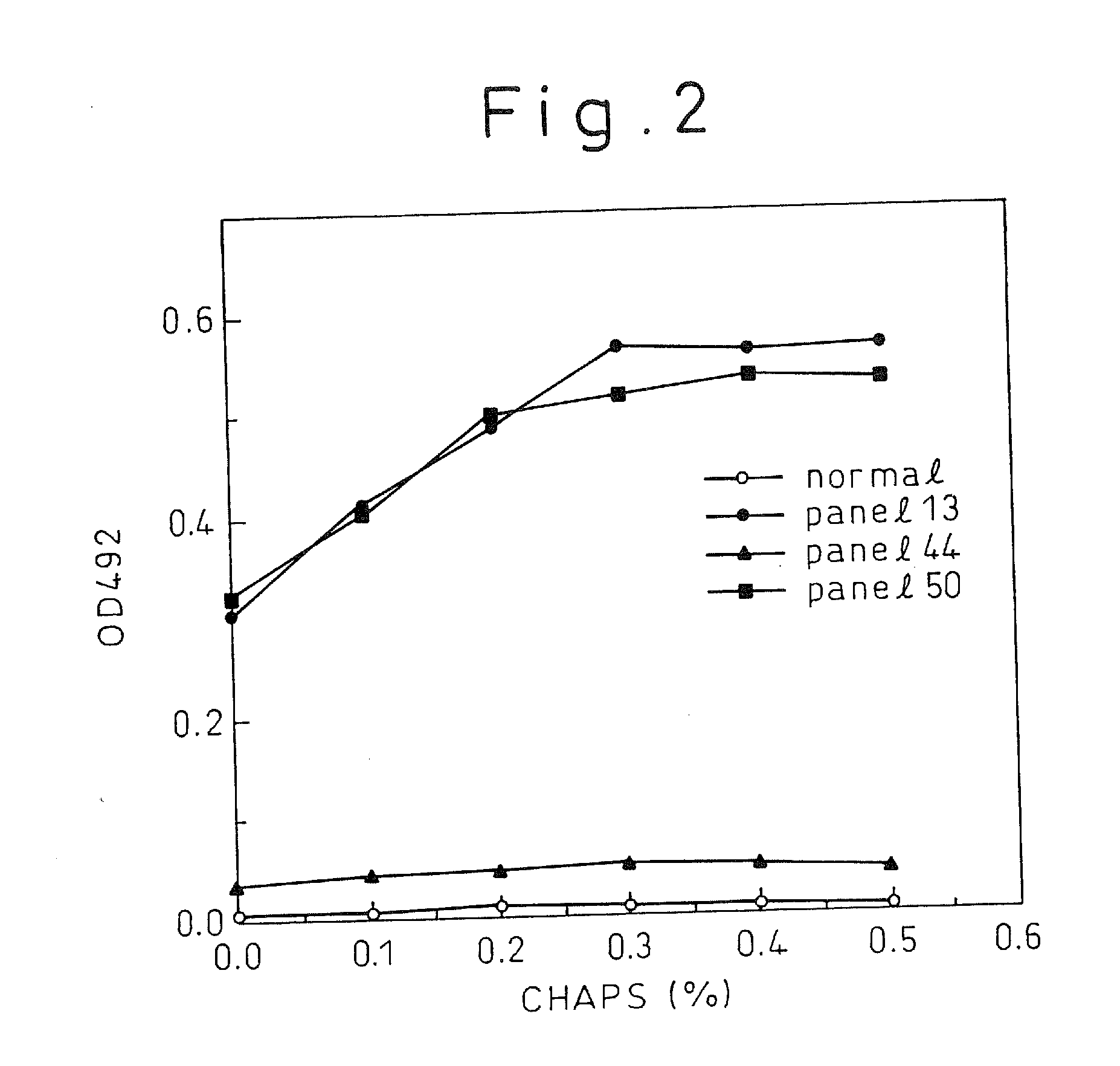
[0137]By an immunoassay using rabbit anti-mouse Ig isotype antibody (manufactured by Zymed), the isotype of each of the monoclonal antibodies C11-14, C11-11, C11-10, C11-7; and C11-3 produced from the above five hybridomas, respectively, was found to be IgG2 for C11-10 and C11-7; and IgGl for CH11-11, C11-14, and C11-3. For the five monoclonal antibodies obtained, epitope analysis was conducted using the synthetic peptides composed of 20 amino acids synthesized according to the sequence derived from the HCV core region. The result indicated, as shown in Table 1, that they were the monoclonal antibodies that specifically recognize part of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com