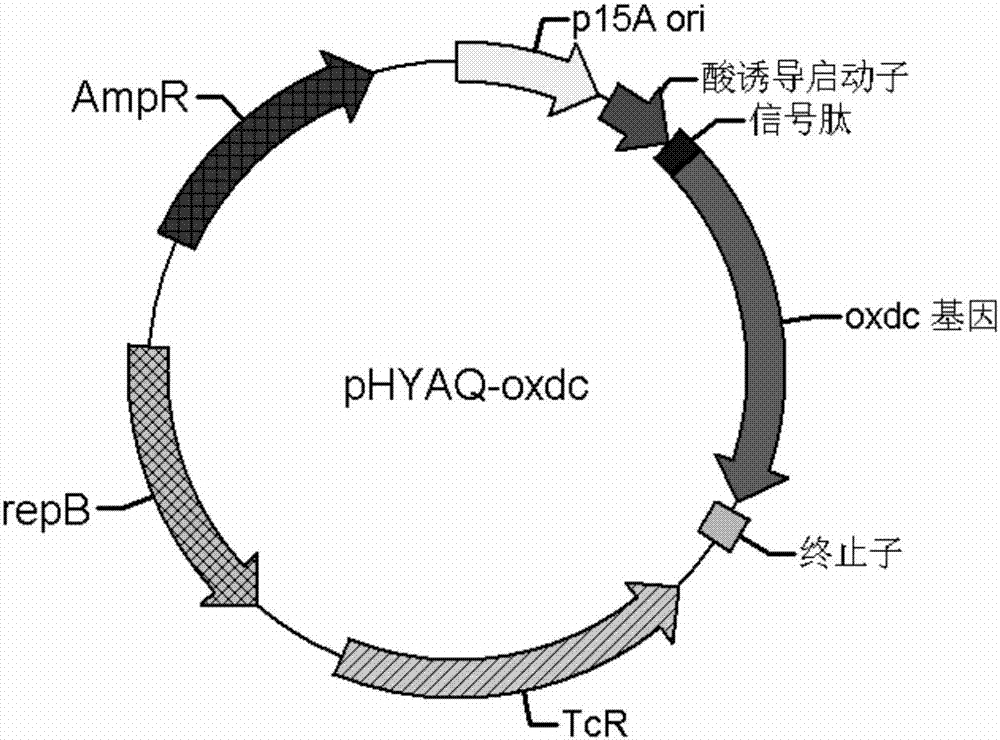
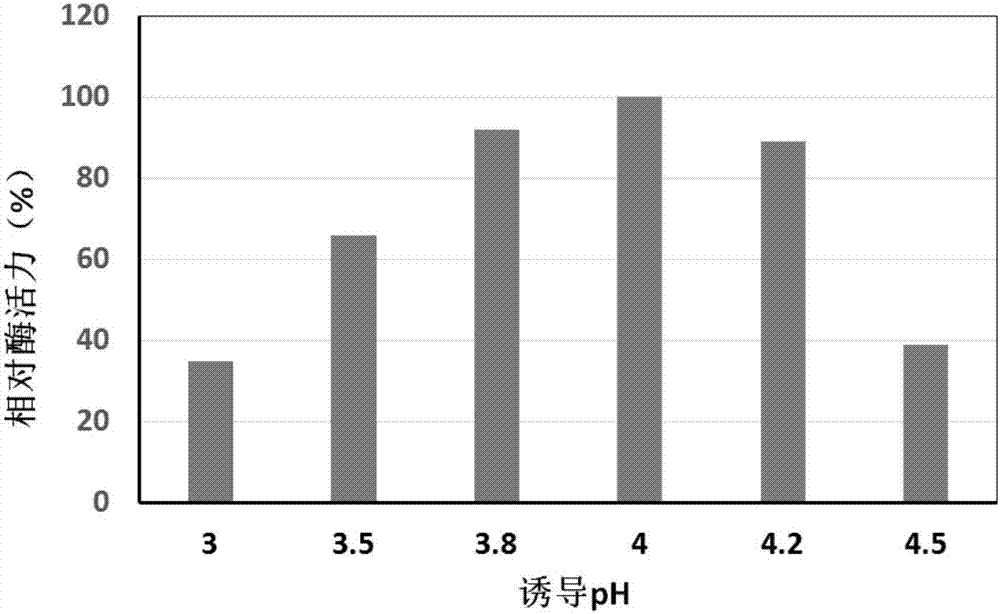
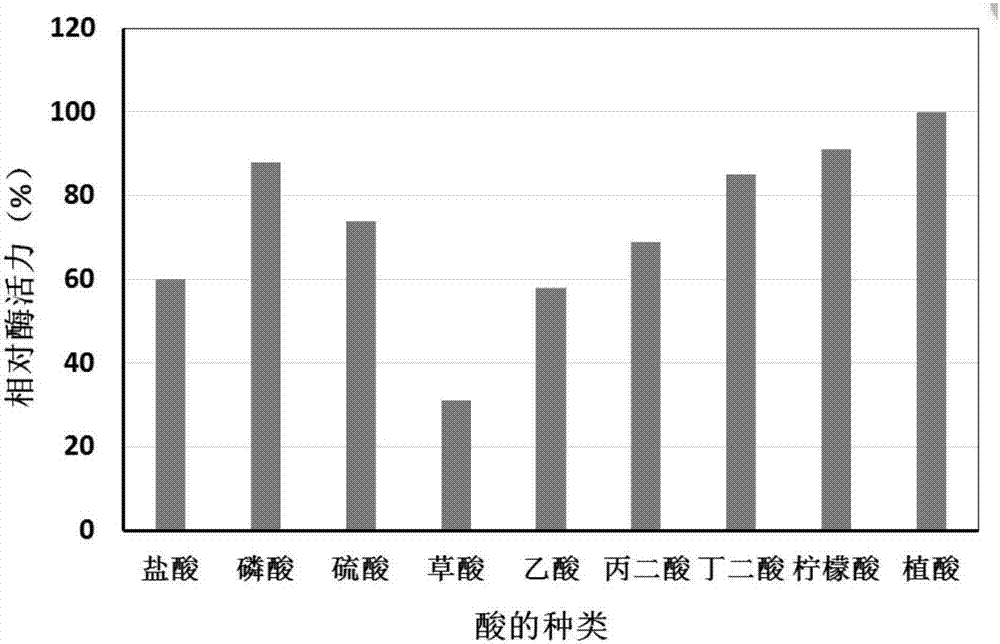
Preparation method and application of oxalate decarboxylase
A technology of oxalate decarboxylase and acid induction, which is applied in the field of preparation of oxalate decarboxylase, can solve the problems such as the inability to efficiently secrete and express OXDC, and achieve the effects of simplifying production and purification steps, reducing production costs, and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The screening of embodiment 1 acid fast bacillus
[0035] The inventors have successively screened more than a hundred species of Bacillus species preserved in the laboratory and purchased by the strain preservation center, including Bacillus circulans CICC10353, CICC10403, CICC21250 and CICC23053, Geobacillus stearothermophilus CICC10142, CICC10267, CICC10362, CICC10392, CICC10425, CICC10782 and CICC10842, etc., Bacillus amyloliquefaciens CICC10025, CICC10035, CICC10036, CICC10063, CICC10074, CICC 10081 and CICC10079, CICC10163, etc. Several strains of Bacillus with strong acid resistance were screened. The screening method is as follows:
[0036] The seed medium is: peptone 10g / L, yeast powder 5g / L, NaCl 5g / L, KH 2 PO 4 2g / L, glucose 10g / L, pH 6.0; screening medium (1L): 20g corn flour, 5g corn steep liquor, 2g yeast powder, 1.0g ammonium chloride, 0.5g manganese chloride, MgSO 4 ·7H 2 O 0.91g, CaCl 2 0.5g, 500mL of citric acid-sodium citrate buffer solution (...
Embodiment 2
[0038] Cloning of embodiment 2 oxalate decarboxylase gene
[0039] The fungal oxalate decarboxylase gene in the present invention is based on the fungal oxdc gene registered in the public database, optimized according to the codons of Bacillus amyloliquefaciens to obtain an optimized gene, and then synthesized through a commercial gene sequencing and synthesis company. The following are several examples of oxalate decarboxylase genes without the original signal peptide obtained through experimental verification based on the codon optimization of fungal-derived oxdc genes registered in the public database (microbial name: database name: original gene accession number: optimized The sequence number of the gene sequence in the sequence table) is as follows:
[0040] Hypsizygus marmoreus: GenBank: LUEZ01000002.1: SEQ ID NO.1;
[0041] Coprinopsis cinerea okayama: GenBank: XM_001836586.2: SEQ ID NO.2;
[0042] Trametes versicolor: GenBank: XM_008041462.1: SEQ ID NO.3;
[0043] H...
Embodiment 3
[0061] Cloning of Example 3 Acid Inducible Promoter
[0062] As the acid-inducible promoter, a promoter derived from Bacillus can be used. In principle, any acid-inducible promoter derived from Bacillus identified so far can be used. Here, an acid-inducible promoter derived from Bacillus subtilis is used as an example. Chromosomal DNA was first obtained from Bacillus subtilis Bs168 strain by conventional methods. Refer to the sequence before the Bacillus subtilis oxdc gene (Gene ID 938620) on the database, design primers F2 and R2 for specifically amplifying the acid-induced promoter region, use the chromosomal DNA of the Bacillus subtilis Bs168 strain as a template, and use the designed Primers (F2 and R2) were used for PCR, thereby obtaining a DNA fragment of the acid-inducible promoter Poxdc, the sequence of which is shown in SEQ ID NO.6.
[0063] F2 (SEQ ID NO.13): 5'-AGCTTGACCACTTCATTTTTC-3'
[0064] R2 (SEQ ID NO.14): 5'-GAAATGTTTCTCTCCTTACAT-3'
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com