Deacetylase capable of producing high-yield glucosamine and coding gene thereof
A technology of glucosamine and deacetylase, which is applied in the field of high-yield glucosamine deacetylase and its coding gene, can solve environmental problems, long production cycle of microbial fermentation method, high equipment requirements and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
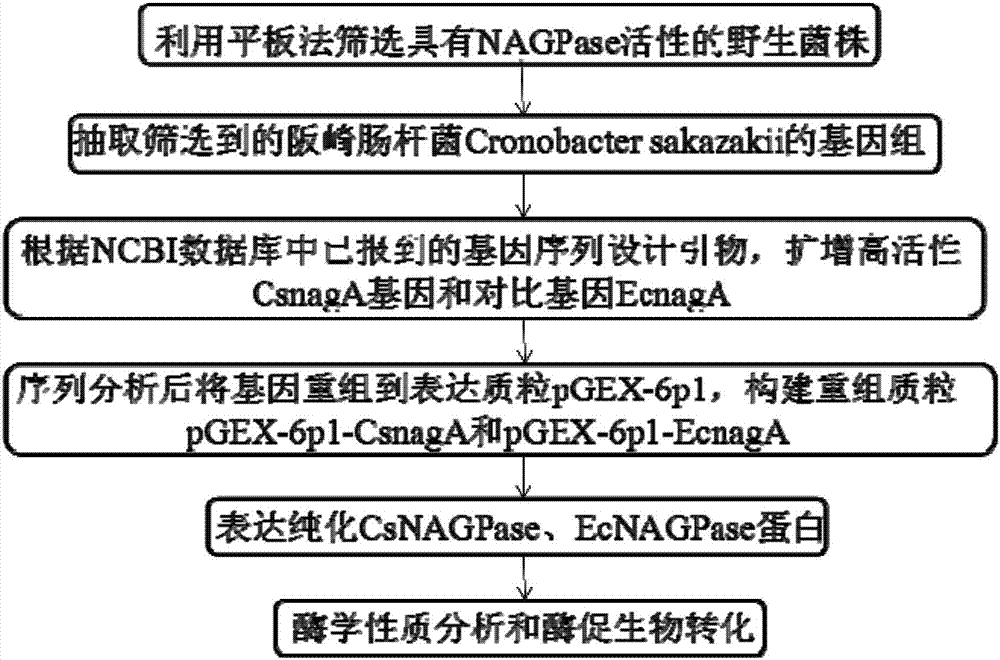
[0032] Example 1 Screening and 16sDNA identification of the natural strain Enterobacter sakazakii producing deacetylase
[0033] After activation on a solid LB (1% peptone, 0.5% yeast powder and 1% sodium chloride, 0.4% agar, pH 7.0) plate, pick a single colony on the enzyme-producing medium (1% peptone, 0.5% Yeast powder, 1% sodium chloride (NaCl), 3% acetylglucosamine, 0.5% phenol red, 0.4% agar, pH 7.0) plate, after 24 hours of cultivation at 37 ° C, pick and produce a large yellow transparent circle strains. After comparative tests, the State Key Laboratory of Agricultural Microbiology of Huazhong Agricultural University, where the applicant works, screened a strain of Enterobacter sakazakii isolated from Xinjiang soil and identified as Enterobacter sakazakii by 16sDNA in 2014. The applicant named the isolated strain Enterobacter sakazakii 0360, Cronobacter sakazakii 0360, and it was sent to China. Wuhan. Wuhan University, China Type Culture Collection Center for preserva...
Embodiment 2
[0041] Cloning of embodiment 2 deacetylase gene
[0042] 1. Extract the chromosomal DNA of Enterobacter sakazakii:
[0043] (1) After the Enterobacter sakazakii strain screened in Example 1 was cultured on a LB liquid medium at 37° C. on a shaker for 12 hours, take 1.5 ml of bacterial liquid in a sterilized Ep tube and centrifuge on a centrifuge at 12,000 rpm 1 minute, discard the supernatant, and collect the bacteria;
[0044] (2) Wash the bacteria twice with TE buffer (50mmol / LTris-HCl, pH8.0; 10mmol / L EDTA, pH8.0), then add 50μl 100μg / ml lysozyme (purchased from sigma company) to suspend the bacteria , 37 ℃ water bath for one hour, so that the lysozyme can fully play its role;
[0045] (3) Add 2 times the total volume of DNA lysate (20mmol / L EDTA, 100mmol / L Tris-Cl (pH=8.0), 2% SDS), mix by inverting several times, and bathe in water at 65°C for 10 minutes;
[0046] (4) Place in -4°C refrigerator for 10 minutes (DNA renaturation)
[0047] (5) Add 150 μL of 5M NaCl, mix ...
Embodiment 3
[0081] Example 3 Deacetylase expression and purification
[0082] 1. Induced expression of recombinant deacetylase in Escherichia coli:
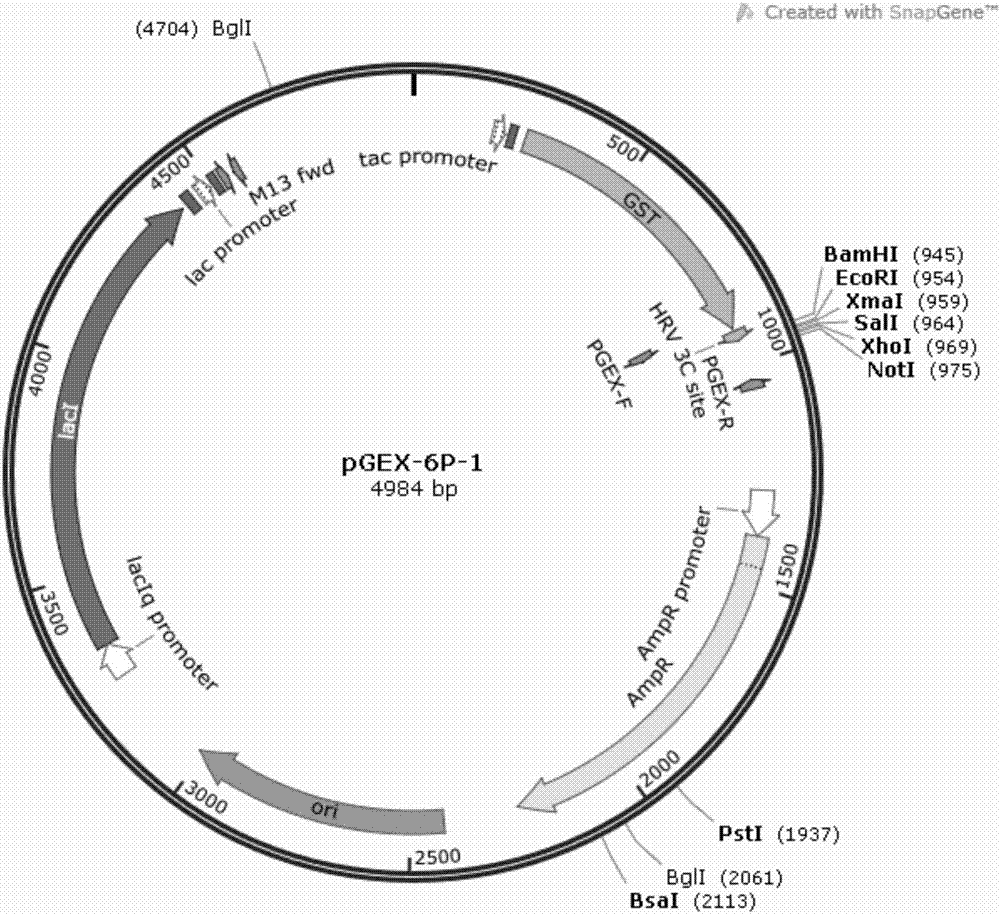
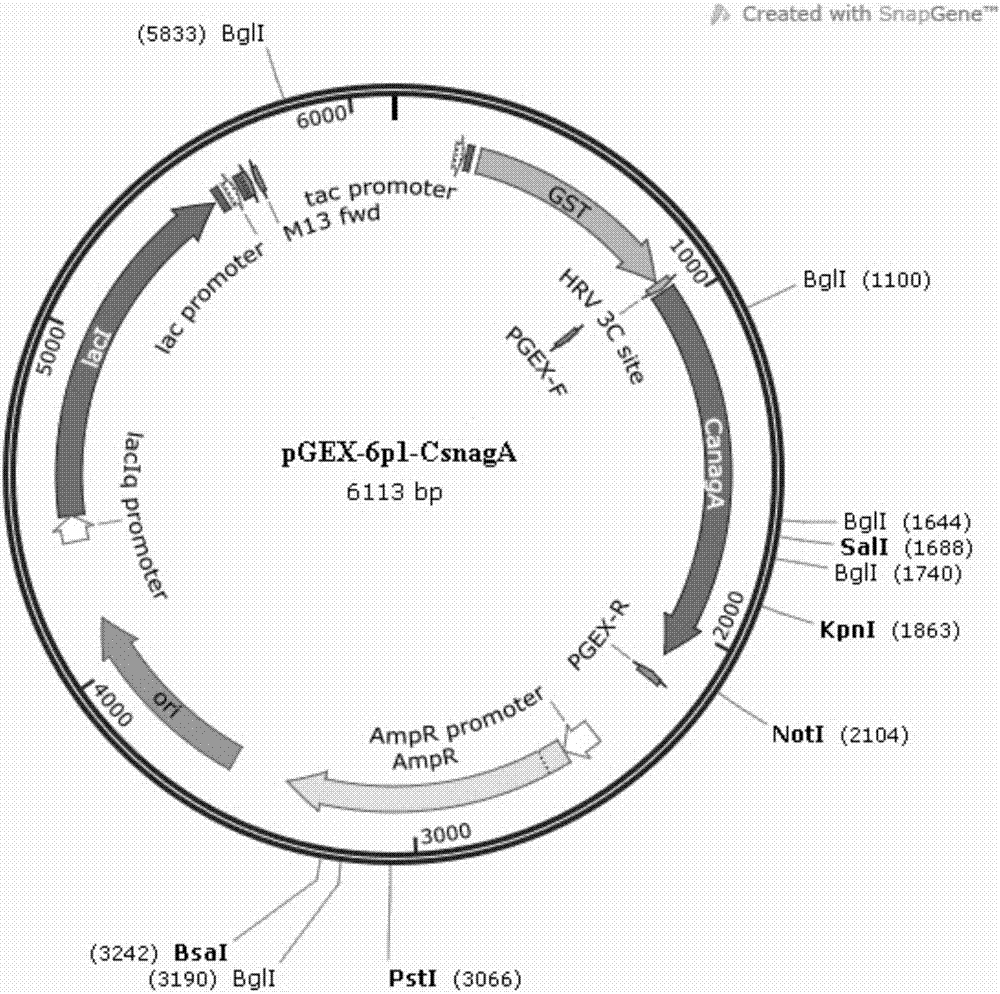
[0083] The recombinant plasmid pGEX-6p1-CsnagA was electrotransformed into E.coli BL21(DE3) for expression and purification, and the transformation steps refer to Example 2. Inoculate the obtained transformants in 100 mL liquid LB medium with 100 μg / mL ampicillin, culture overnight at 37°C, transfer to 1L LB liquid medium with 100 μg / ml ampicillin at 37°C Cultivate for 2-3hrs to make OD 600 When it reaches 0.6-0.7, add isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM, and incubate at 18° C. on a shaker at 180 rpm for 14 hours. Centrifuge at low temperature (4°C) and high speed (8000r / min) to collect the bacterial cells, then centrifuge at 8000r / min for 8min, and then use an appropriate amount of ddH 2 O resuspended bacterium, centrifuged at 8000r / min for 8min, then phosphate buffered saline, namely PBS (prepar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com