Engineering probiotic capable of degrading uric acid in intestinal tract, and preparation method and application of engineering probiotic
A kind of probiotics and engineering technology, applied in the field of probiotics preparation, can solve the problems of no practical application value, no expression efficiency of uric acid permeation enzyme gene, no discovery of natural probiotics to efficiently absorb exogenous uric acid, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1 Obtaining of urate oxidase coding sequence
[0064] Bacillus subtilis urate oxidase (GenBank: WP_003242600.1), Candida utilis urate oxidase (GenBank: P78609.1), Aspergillus niger urate oxidase (GenBank: XP_001399739.1), Trichoderma reesei urate oxidase (GenBank: XP_006963697.1), Cordyceps militaris urate oxidase (GenBank: XP_006674629.1), and Kluyveromyces marxense urate oxidase (GenBank: XP_022674397.1) coding sequences were extracted from the genomic DNA of the corresponding species. The primers in Table 1 amplified the full-length sequence of urate oxidase, and after sequence comparison and analysis, the sequence was spliced after manually removing introns from eukaryotic genes.
[0065] Aspergillus flavus urate oxidase (GenBank: XP_001826198.1), Arthrobacter globosa urate oxidase (GenBank: D0VWQ1.1), Microbacterium urate oxidase (GenBank: AEY68606.1), porcine urate oxidase (GenBank: NP_999435.1 ) sequence was synthesized by Wuhan Tianyi Huiyuan Biotech...
Embodiment 2
[0069] The construction of the Lactobacillus casei engineering bacterium of embodiment 2 recombinant expression uric acid permeation enzyme
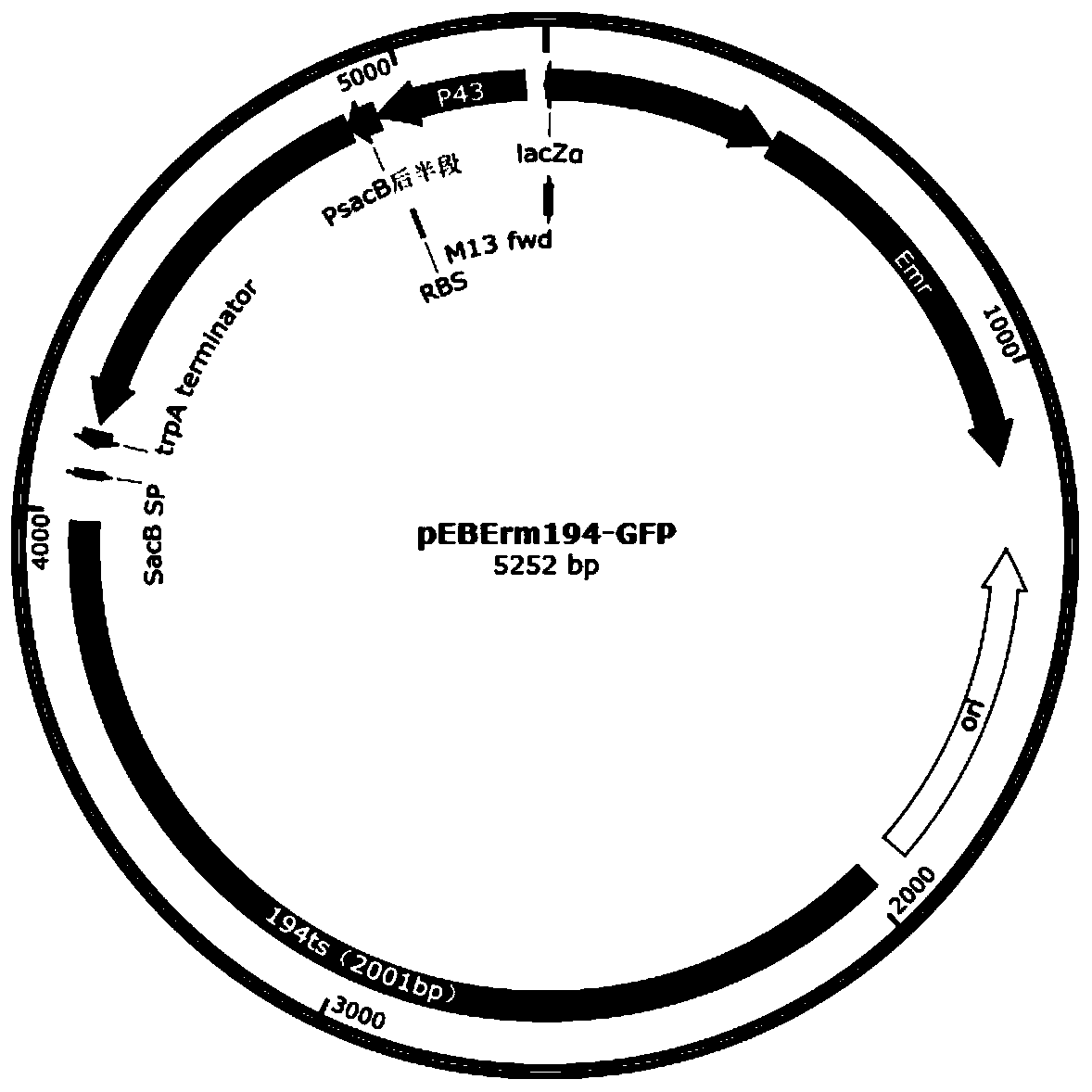
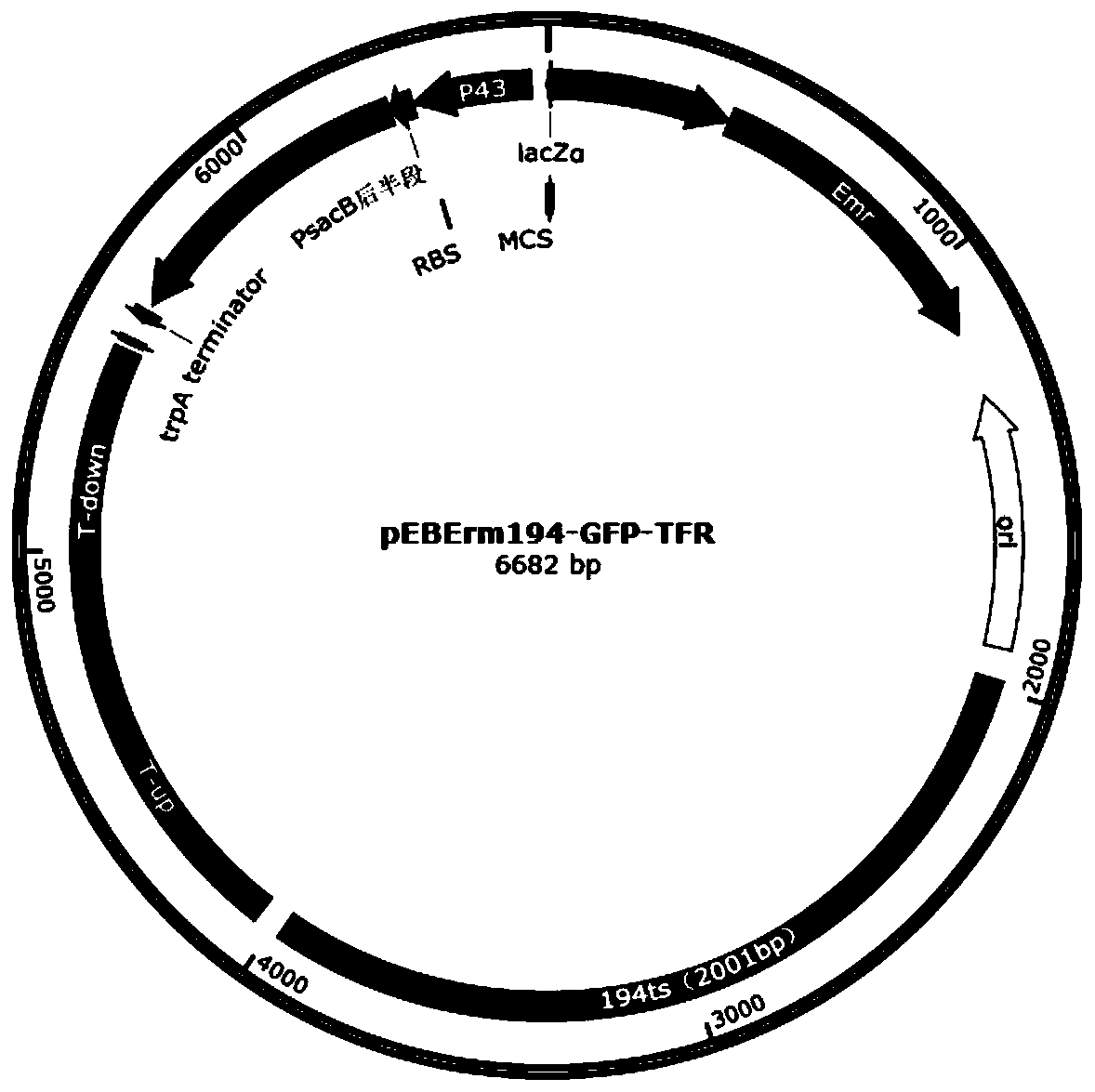
[0070] 2.1pEBErm194-GFP-TFR vector construction
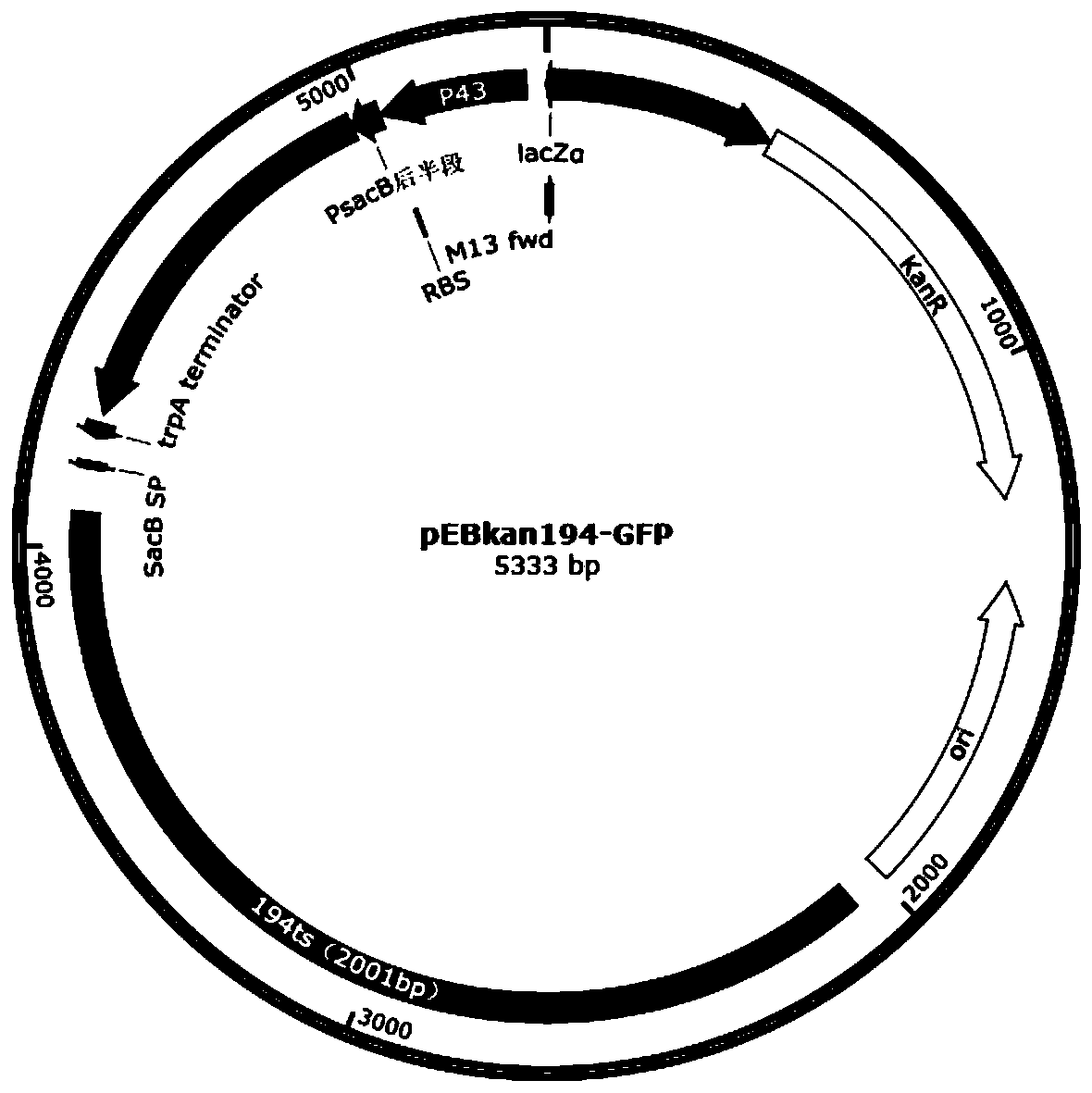
[0071] Using Lactobacillus casei ATCC393 genomic DNA as a template, using the primers TF-up-F / TF-up-R, TF-down-F / TF-down-R in Table 2, PCR amplification of the upstream homology arm TF-up and the downstream homology arm TF-down, and then use TF-up-F / TF-down-R as primers to amplify TF-up+down by overlapping PCR, and recover the PCR fragment with a DNA agarose gel recovery kit; The pEBErm194-GFP plasmid was used as a template, and the primers pEBErm194-F / pEBErm194-R in Table 2 were used to amplify a vector backbone fragment of about 5300 bp by PCR, digested with DpnI for 2 hours, and recovered the vector fragment with a DNA agarose gel recovery kit; The digested and recovered vector backbone and up+down overlapping fragments were assembled according to the method of the seamless cloning ki...
Embodiment 3
[0084] The construction of the Lactobacillus casei engineering bacterium of embodiment 3 recombinant expressing uric acid oxidase
[0085] 3.1 Construction of urate oxidase integration vector
[0086] Using the HCE promoter synthesized from the whole gene of the database sequence (GenBank: DJ022754.1) as a template, use the primers HCE-F / HCE-R in Table 4 to amplify the strong constitutive promoter HCE promoter (SEQ No.20) by PCR; Using the CU-UOX gene sequence as a template, use the primers CUUOX-F / CUUOX-R in Table 4 to amplify the CU-UOX gene sequence by PCR; use the rrnBT1T2 synthesized by the whole gene as a template, and use the primers rrnBT1T2-F / rrnBT1T2-R, PCR amplification terminator rrnBT1T2 (SEQNo.21). Then use these three fragments as templates, use primers HCE-F / rrnBT1T2-R, overlap PCR to amplify HCE promoter+CUUOX+rrnBT1T2, and use DNA agarose gel recovery kit to recover PCR fragments. Using the plasmid pEBErm194-GFP-TFR as a template, using the primers P-down-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com