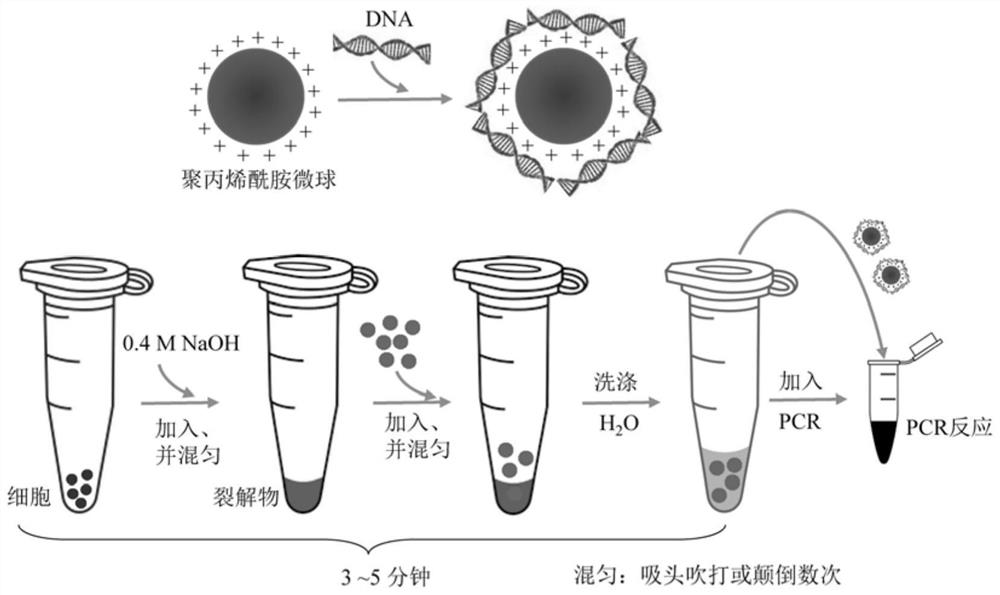
A kind of polyacrylamide microsphere for rapid extraction of nucleic acid and its preparation method and application
A technology of polyacrylamide microspheres and polyacrylamide, which is applied in the field of nucleic acid extraction and detection of biology, and can solve the problems of inability to improve machine automation, low DNA absorption and elution efficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
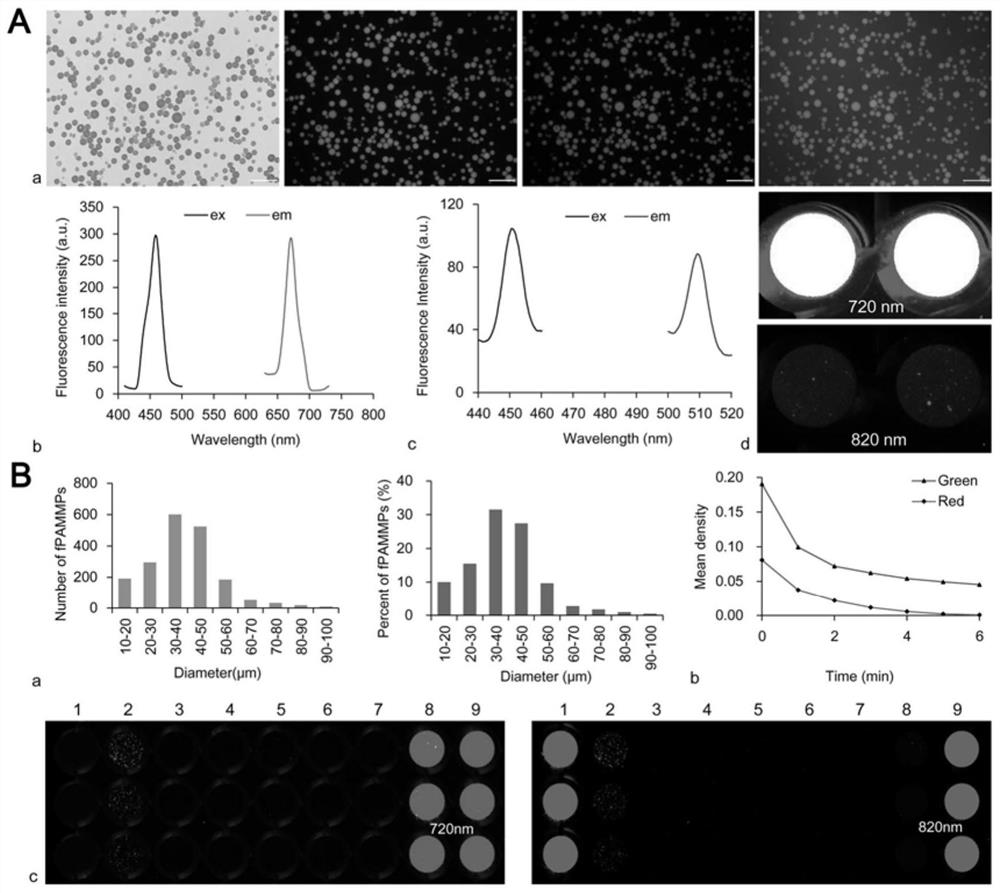
[0052] Synthesis and characterization of embodiment 1 fPAMMP
[0053] method:
[0054] 1. Materials: acrylamide (AM), N-(3-aminopropyl)methacrylamide hydrochloride (APMA) were purchased from SigmaAldrich (USA). Glutaraldehyde (GTA), Span 80 and NaOH were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Mineral oil, ammonium persulfate (APS), N,N,N',N'-tetramethylethane-1,2-diamine (TEMED) and N,N'-methylenebisacrylamide (MBA) Purchased from Biosharp (Hefei, China). Polylysine (ε-PL) was purchased from Shanghai Shifeng Biotechnology Co., Ltd. (Shanghai, China). 2×Premix PrimerStar HS (product number: R040A) was purchased from Takara (Dalian). qPCR Master Mix (2x) was purchased from Promega. Oligonucleotides were produced by Sangon Biotech.
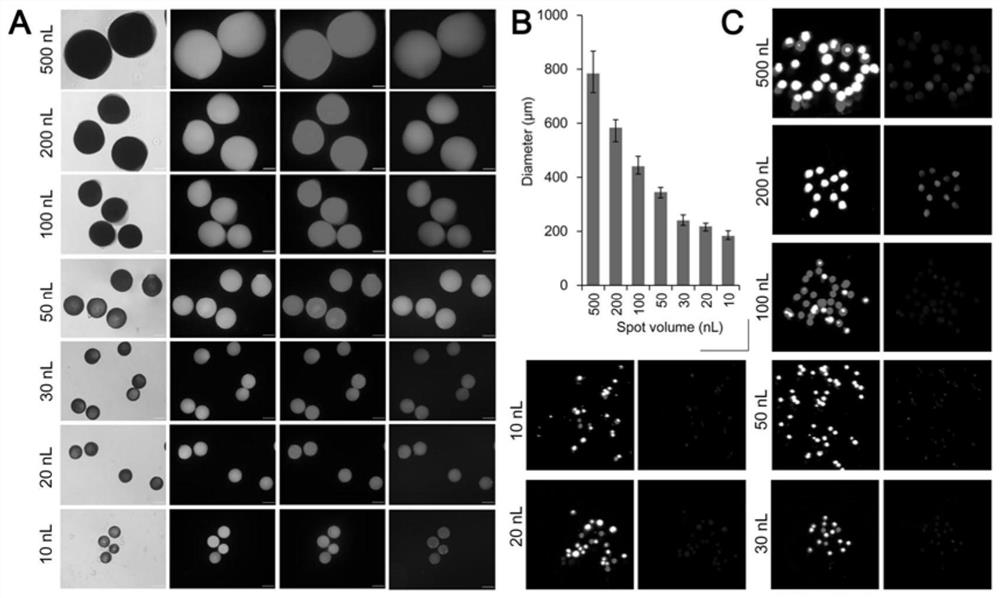
[0055] 2. Synthesis of fPAMMP by spot polymerization: The preparation scheme of PAMMP is as follows: 264mg AM, 80mg MBA and 12mg APMA were dissolved in 1mL deionized (DI) water under ultrasonic to obtain a unifo...
Embodiment 2
[0062] Example 2 The ability of fPAMMP to capture DNA
[0063] method:
[0064] Mix 2 μg of purified SiHa cell genomic DNA with 80 μL of fPAMMP aqueous solution and invert several times. fPAMMP was then washed to remove excess gDNA. The fPAMMP@SiHa DNA was detected by agarose gel electrophoresis.
[0065] result:
[0066] In order to develop a fast and simple DNA extraction method, the present invention first investigated the ability of fPAMMP to bind to DNA. Genomic DNA from SiHa cells was mixed with fPAMMP and inverted several times. fPAMMP was then washed to remove excess gDNA. Gel electrophoresis results show that fPAMMP can effectively bind or at least capture DNA within seconds due to its positive charge ( Figure 7 A, a). Negatively charged DNA binds to fPAMMP through electrostatic interactions. Due to the microscale size of fPAMMP, it cannot pass through the gel, so the positive results of DNA staining are shown in the gel wells, indicating that the DNA is firml...
Embodiment 3
[0067] Example 3 Carry out DNA extraction and direct PCR amplification with fPAMMP
[0068] method:
[0069] 1. DNA extraction with fPAMMP
[0070] Bacterial DNA extraction: Escherichia coli BL21 and DH5α in glycerol stocks were streaked onto Luria Bertani (LB) agar plates and incubated at 37°C for 16 hours, respectively. A single colony of Escherichia coli that was well separated was inoculated into a test tube containing 2 mL of LB liquid medium, and incubated at 37 °C for 8 hours with vigorous shaking at 220 rpm. The culture was then centrifuged at 12000 rpm (5415D, Eppendorf) for 5 minutes and the supernatant was discarded. Resuspend the obtained pellet in 200 µL of deionized water and mix with an equal volume of 0.4 M NaOH. Cells were lysed by gentle inversion several times. Then 100 μL fPAMMP aqueous solution was added to the mixture and incubated in a rotator at room temperature for 5-30 minutes. The fPAMMP (fPAMMP@DNA) that had captured bacterial genomic DNA was c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com