Expression and optimization method of novel arabinoside enzyme gene
A technology of arabinosidase and expression method, which is applied in the fields of enzymology, bioinformatics, genetic engineering, and molecular biology, and can solve the problems of poor utilization rate, single type of arabinosidase, and low production level, so as to improve the expression level , wide temperature and pH adaptability, high activity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Directed mutagenesis of arabinosidase gene
[0029] 1. Subcloning of the Arabinosidase Gene
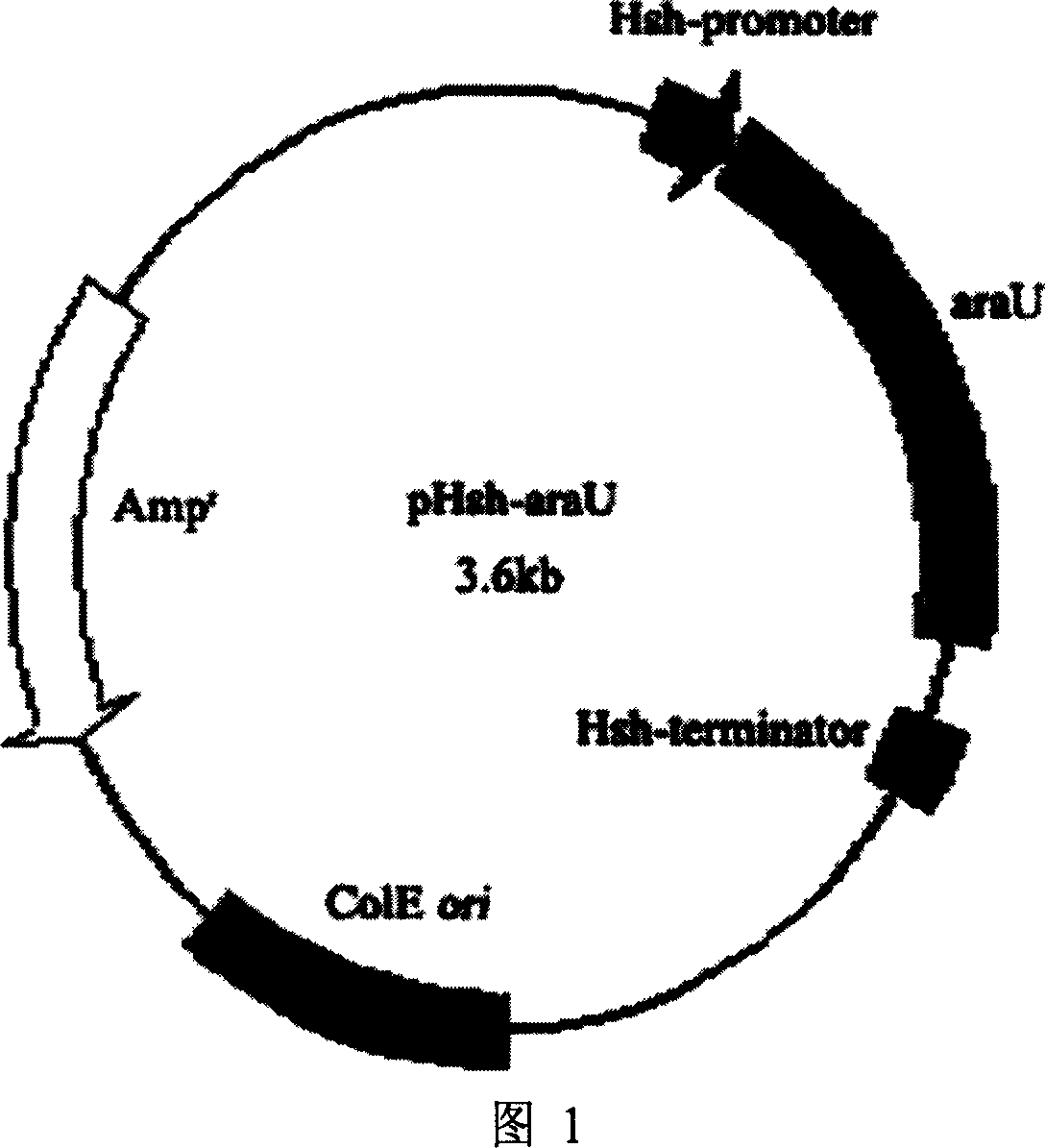
[0030] Primers were designed according to the sequence of the arabinosidase gene (GenBank accession number DQ324528), upstream 5'-CCC CT GCAG Add Pst I site in front of A GAATGG CACGG TCAAA GTC-3'; downstream 5'-CCC AA GCTT ATTAAG CCAGA TCGAC GTCAA AC-3' introduces a Hind III site. Two stop codons UAA, UAA were added to enforce translation termination. Using the gene araB as a template, PCR amplification was carried out with synthetic primers. The amplification conditions were 95°C, 5min; the timing was paused, DNA polymerase Pyrobest was added, and 40 μL paraffin oil was added to seal; 35 cycles (94°C, 50s; 51 ℃, 90s; 72℃, 3min); 72℃, 10min; the reaction was stopped, and kept at 4℃. The amplified product was double-digested with Pst I and Hind III, and connected with the expression vector pHsh double-digested with the same enzyme to obtain the recombinant pla...
Embodiment 2
[0033] Embodiment two: the expression of arabinosidase gene and mutant thereof
[0034] The cultivation of recombinant bacteria: recombinant plasmids pHsh-araB, pHsh-ara, pHsh-araU; the pHsh-araB, pHsh-ara, pHsh-araU plasmids were transferred into Escherichia coli strain JM109 by electroporation (electroporation), and selected Inoculate 20ml of LB medium containing 100μg ml ampicillin into 20ml of LB medium containing 100μg ml of ampicillin, carry out shaking culture in a shaker at 37°C to the logarithmic phase, and transfer to 300ml of LB medium containing 100μg ml of ampicillin at a 2% inoculum size , in a 200rpm 30°C shaker for shaking culture to OD 600 After heat shock induction at 42°C for 8 hours at about 0.6, the cells were harvested by centrifugation.
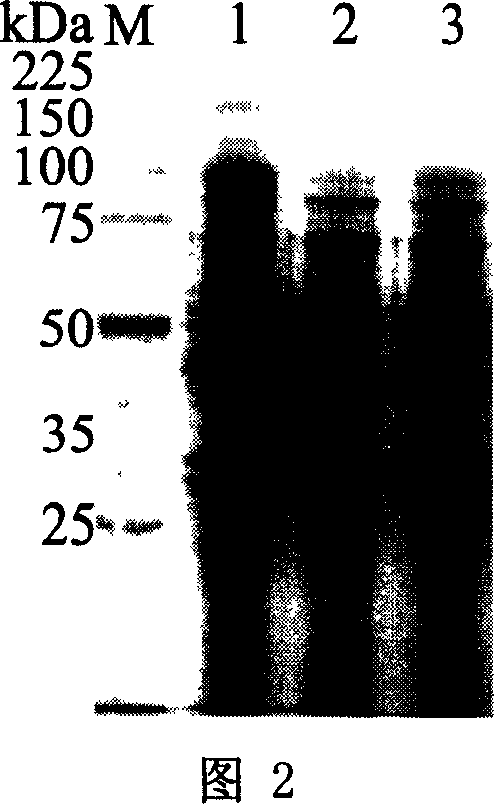
[0035] Preparation of recombinant arabinosidase: suspend cells in phosphate buffer, and lyse the cells with a high-pressure cell disruptor; fractionally precipitate the cell lysate with 50%-80% ammonium sulfate to obt...
Embodiment 3
[0036] Example 3: Enzymatic Properties and Degradation Test of Recombinant Arabinosidase
[0037] Enzymatic analysis was performed on the expression products of araB, ara and araU in Example 2, and their enzymatic properties were the same.
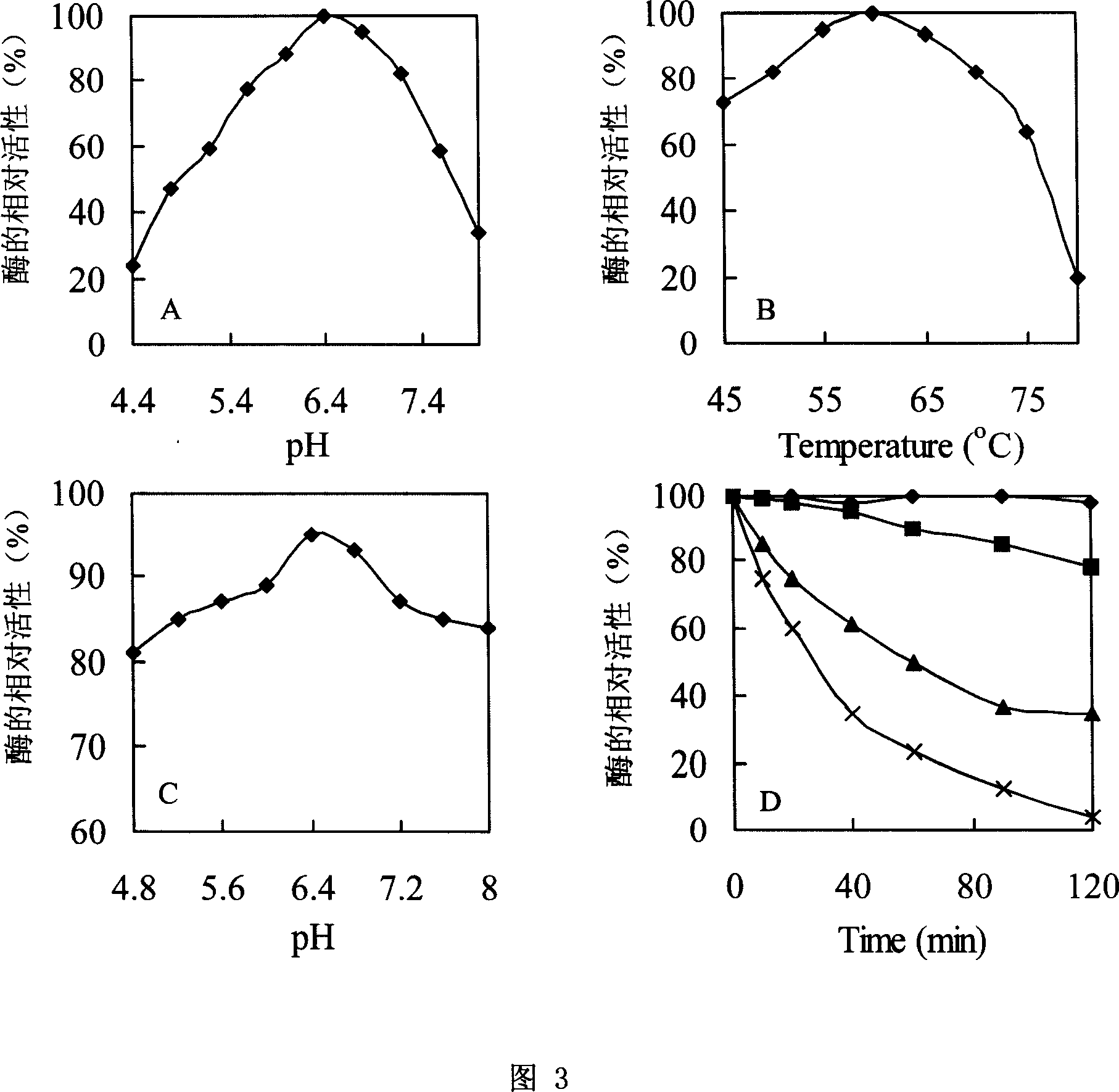
[0038] Determination of the optimum pH: the pure enzyme of the recombinant enzyme was measured in 50mmol L imidazole-potassium hydrogen phthalate buffer (IP buffer) at pH 44-8.0, and the enzyme activity was measured at 60°C. Calculate the relative enzyme activity to determine the optimum pH. As a result, the optimal pH of the enzyme was 6.4 (Fig. 3A).
[0039] Determination of the optimum reaction temperature: measure the enzyme activity of the recombinant enzyme at 45°C-80°C and pH 60 (IP buffer), and calculate the relative enzyme activity to determine the optimum reaction temperature. The results showed that the optimum reaction temperature was 60°C, but the enzyme maintained a relatively high enzyme activity in the range of 45°C-75°C ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com