Nucleic Acid Mediated Electron Transfer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Synthesis of an Oligonucleotide Duplex with Electron Transfer Moieties at the 5′ Termini
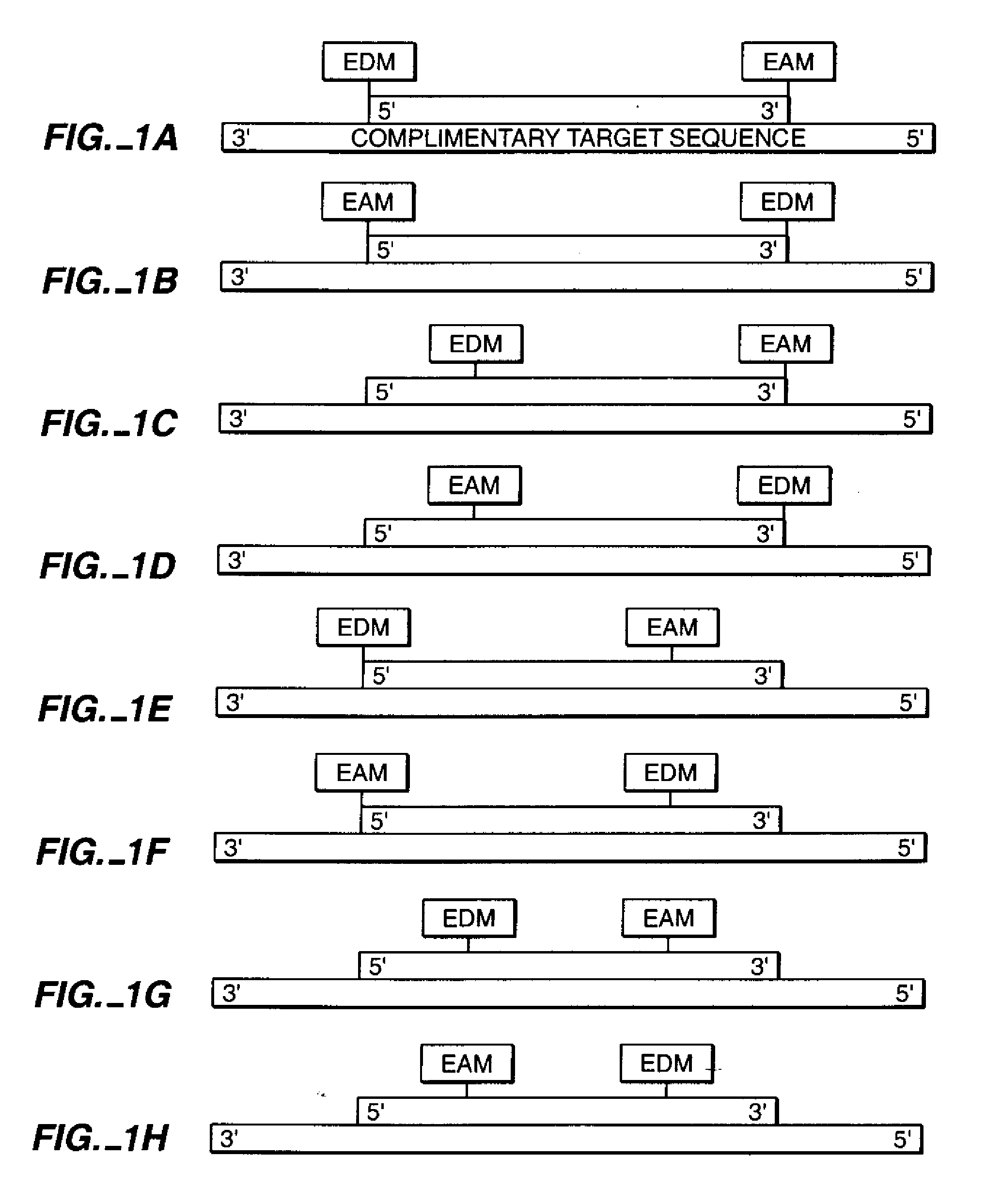
[0174]In this example an eight nucleotide double stranded nucleic acid was produced, with each single strand having a single electron transfer moiety covalently attached to the 5′ terminal uridine nucleotide at the 2′ carbon of the ribose sugar.
Step 1: Synthesis of 5′-di(p-methoxyphenyl)methyl ether-2′-(trifluoroacetamido)-2′-deoxyuridine
[0175]2′-(trifluoroacetamido)-2′-deoxyuridine (2.0 g, 5.9 mmoles) prepared by minor modification of published procedures (Imazawa, supra) was repeatedly dissolved in a minimum of very dry CH3CN and rotary evaporated to dryness and then transferred to inert atmosphere vacuum line and further dried for a period of 1 hour. The following procedure for the synthesis of the material was adapted from Gait (supra): Under positive pressure argon, the material was dissolved in freshly dried and distilled pyridine and with stirring, 0.05 equivalents (wt.) of 4-dimethylamino...
example 2
Synthesis of Long DNA Duplexes with Electron Transfer Moieties at the 5′ Termini
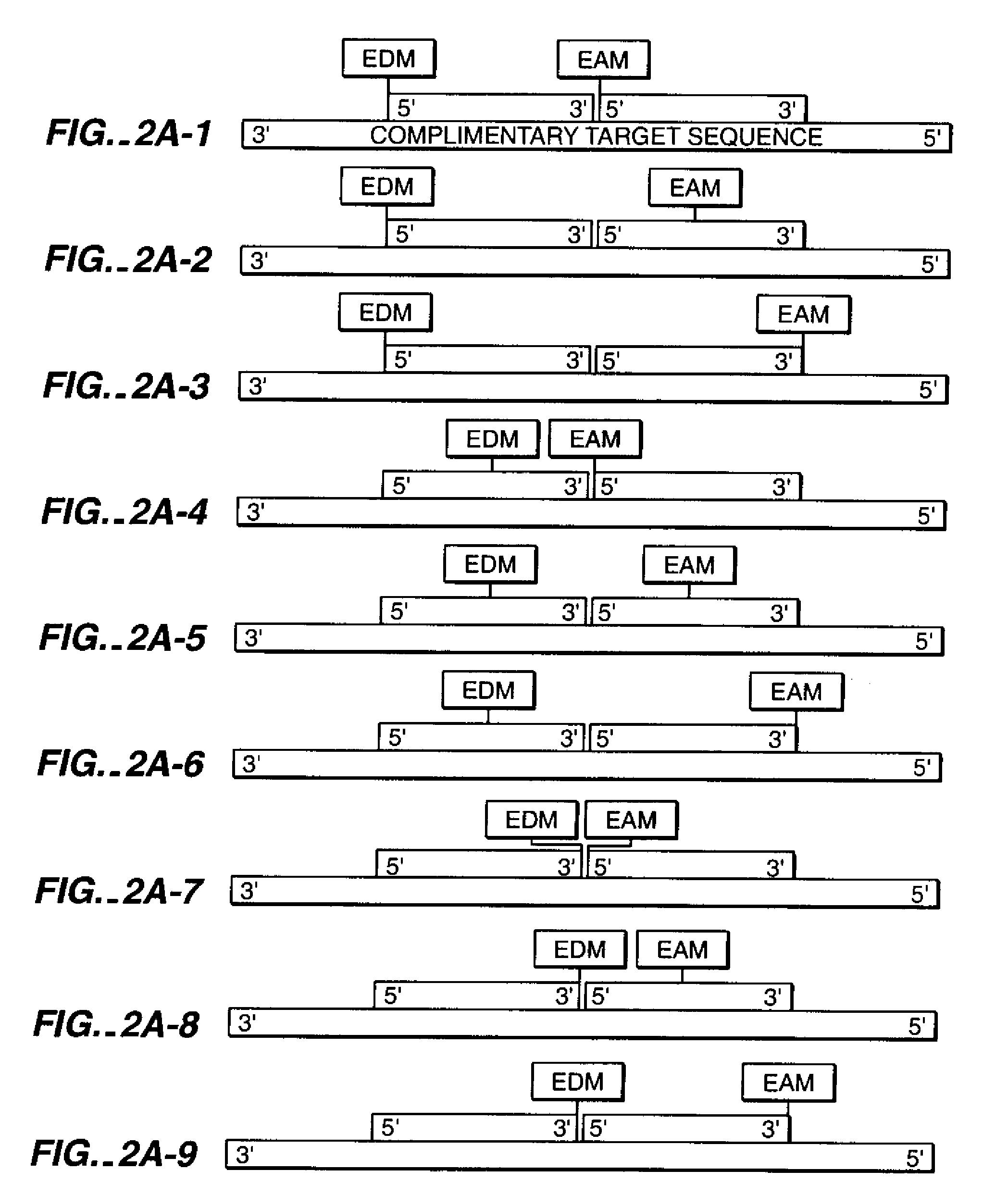
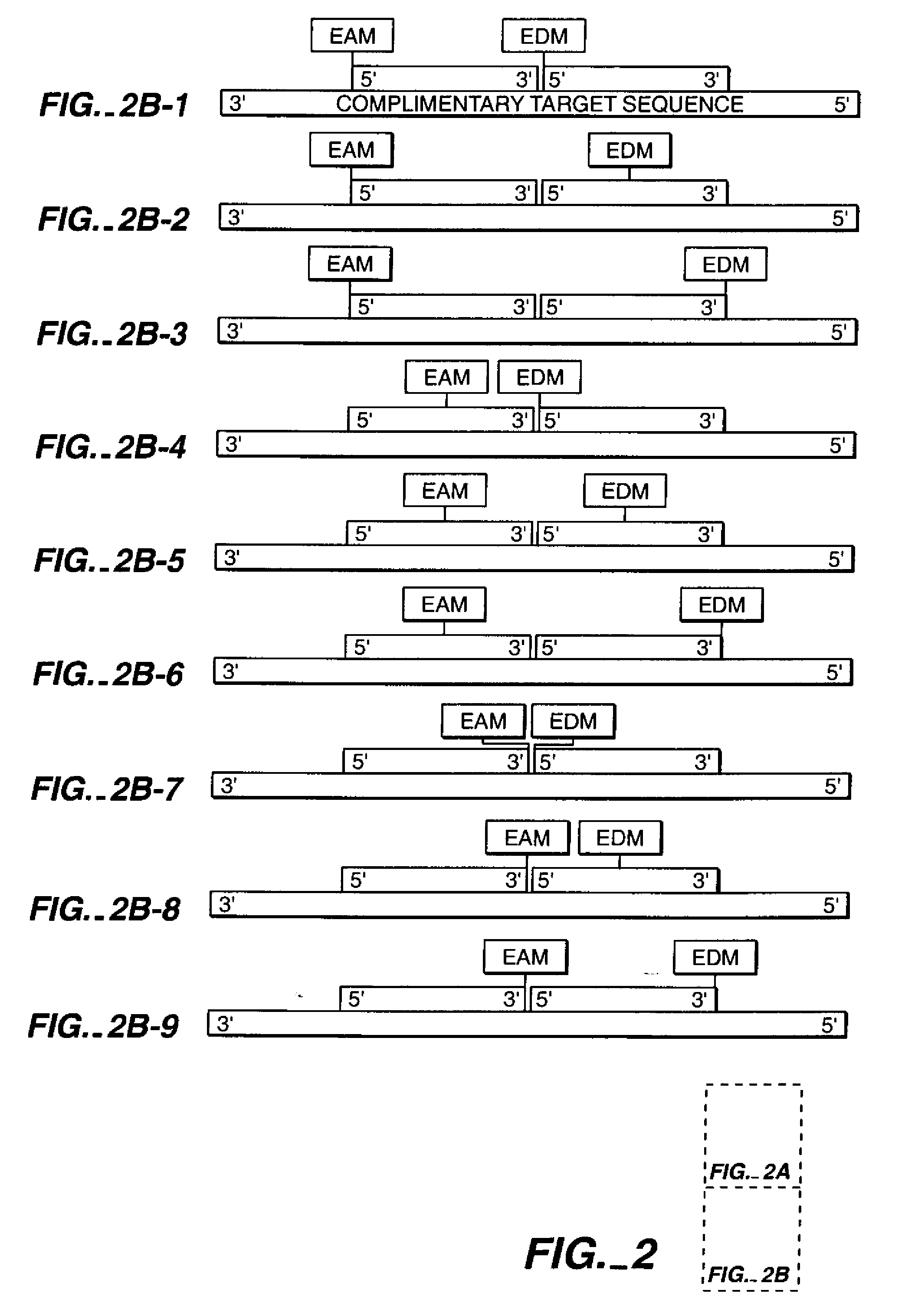
[0179]In this example, an in vitro DNA amplification technique, PCR (reviewed in Abramson et al., Curr. Op. in Biotech. 4:41-47 (1993)) is used to generate modified duplex DNA by polymerization of nucleotides off modified primer strands (Saiki et al., Science 239:487 (1988)). Two oligonucleotides 18 bases in length and not complementary to each other are synthesized with amino-modification to the 2′-ribose position of the 5′ nucleotides, as in example 1.
[0180]A series of oligonucleotides of increasing lengths starting at 40 bases are chemically synthesized using standard chemistry. Each of the PCR templates shares a 5′ sequence identical to one modified 18mer. The 3′ end of the template oligonucleotide shares a sequence complementary to the other 18mer.
[0181]PCR rapidly generates modified duplex DNA by the catalysis of 5′-3′ DNA synthesis off of each of the modified 18mers using the unmodified strand as ...
example 3
Synthesis of Covalently Bound Electron Transfer Moieties at Internucleotide Linkages of Duplex DNA
[0183]In this example, alternative backbones to phosphodiester linkages of oligonucleotides are employed. Functional groups incorporated into these internucleotide linkages serve as the site for covalent attachment of the electron transfer moieties. These alternate internucleotide linkages include, but are not limited to, peptide bonds, phosphoramidate bonds, phosphorothioate bonds, phosphorodithioate bonds and O-methylphosphoramidate bonds.
[0184]The preparation of peptide nucleic acid (PNA) follows literature procedures (See Engholm, supra), with the synthesis of Boc-protected pentafluorophenyl ester of the chosen base (thymidine). The resulting PNA may be prepared employing Merrifield's solid-phase approach (Merrifield, Science, 232:341 (1986)), using a single coupling protocol with 0.1 M of the thiminyl monomer in 30% (v / v) DMF in CH2Cl2. The progress of the reaction is followed by q...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com