Nucleic acid chip, preparation method and application thereof
A nucleic acid chip and nucleic acid technology, applied in biochemical equipment and methods, nucleotide libraries, protein nucleotide libraries, etc., to achieve the effects of low cost, expanded application range, and ease of use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0060] Example 1 cell culture:
[0061] The cells used in the experiment include Hela, HepG2 and U2-OS. The cell culture medium was Dulbecco's Modified Eagle Medium (DMEM, Invitrogen), which contained 10% fetal bovine serum and 1% double antibody. Culture conditions: humidity 95%, temperature 37°C, 5% CO 2 . When the cells are subcultured, wash them with PBS first, then add 0.25% trypsin containing 5mM EDTA, and add serum to terminate the digestion when the edges of the cells are observed to float and appear round. Use a pipette to blow off the adherent cells, centrifuge to remove the supernatant, and obtain the deposited cells. After adding an appropriate amount of medium to dilute, use a cell counting plate to count, and then culture.
example 2
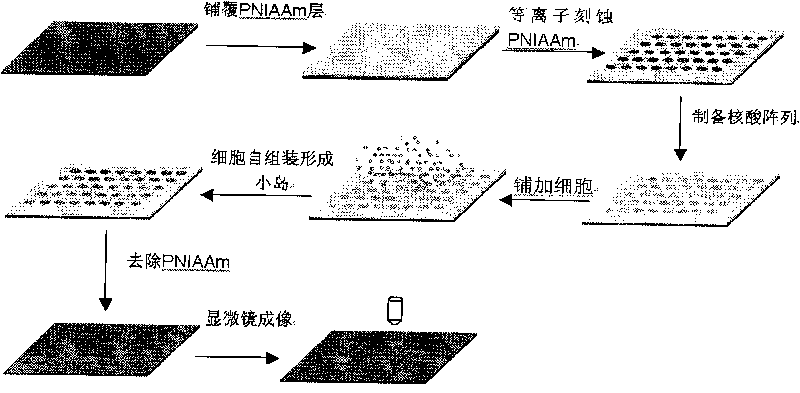
[0062] Example 2 utilizes dispenser to prepare nucleic acid lattice
[0063] Nucleic acid array preparation was performed using a dispenser based on microfluidic technology (Wang, et al. Lap Chip, 2009). According to different experimental requirements, other spotting instruments can also be used. The dispenser is designed with six independent pipes so that six different samples can be dispensed simultaneously. An air pressure pulse (0.04 MPa, 15 msec) generated by compressed air can drive approximately 110 nanoliters of liquid reagent to fill the surface of a well that is not covered by polymer. When spotting, the chip was placed on a 37°C temperature-controlled platform to prevent liquid reagents from dissolving the polymer. After spotting, clean the distributor tube with ultrapure water and dry it with compressed air for next use.
example 3
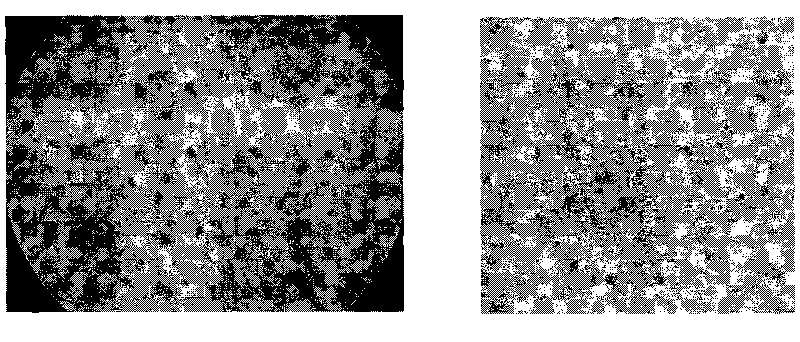
[0064] Example 3 Cell Self-Assembled Small Islands
[0065] Clean the surface of a glass slide (25 mm X 25 mm) commonly used in the laboratory with detergent and ultrapure water. After drying, drop a layer of polymer film on the surface, use 65 microliters of 6% (w / v) Poly(N-isopropylacrylamide) (Sigma or PolySciences) ethanol solution, dry, and store at room temperature for 12 hours. Prepare a silicon wafer mask according to the experimental requirements, and micro-holes of different sizes or shapes can be engraved on it by using micro-processing technology. Specifically, the silicon wafer mask is covered on the glass slide after the film is dried, and etched in an oxygen plasma etching machine. The etching conditions are 50pa oxygen pressure, 200W power, and the etching time is about 3.5 minutes. After the etching is completed, put the glass slide into the ultra-clean bench for ultraviolet irradiation disinfection, and then use the dispenser to process the siRNA microarray...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com