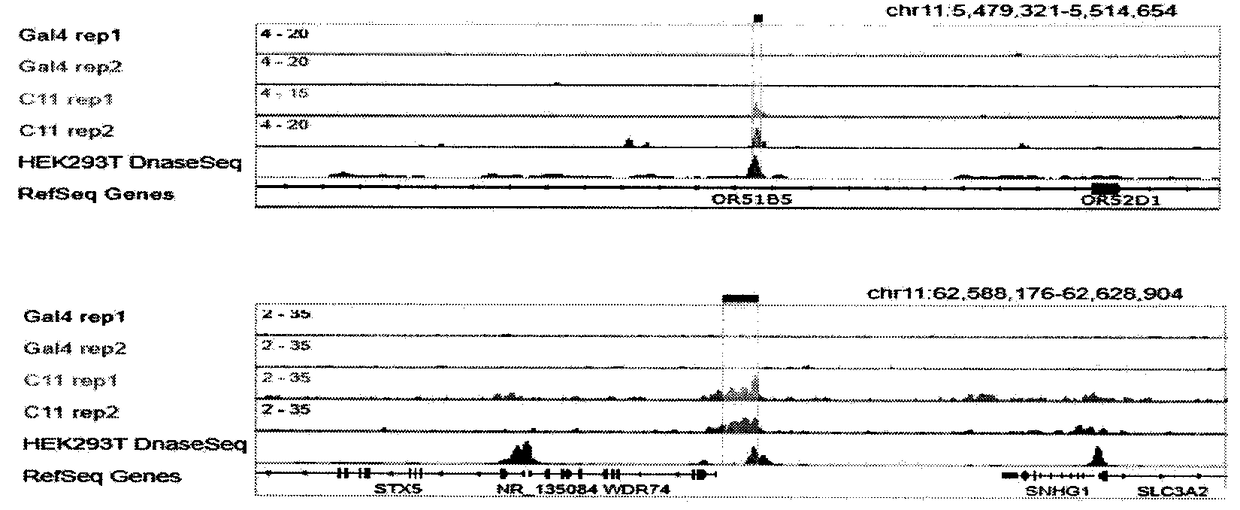
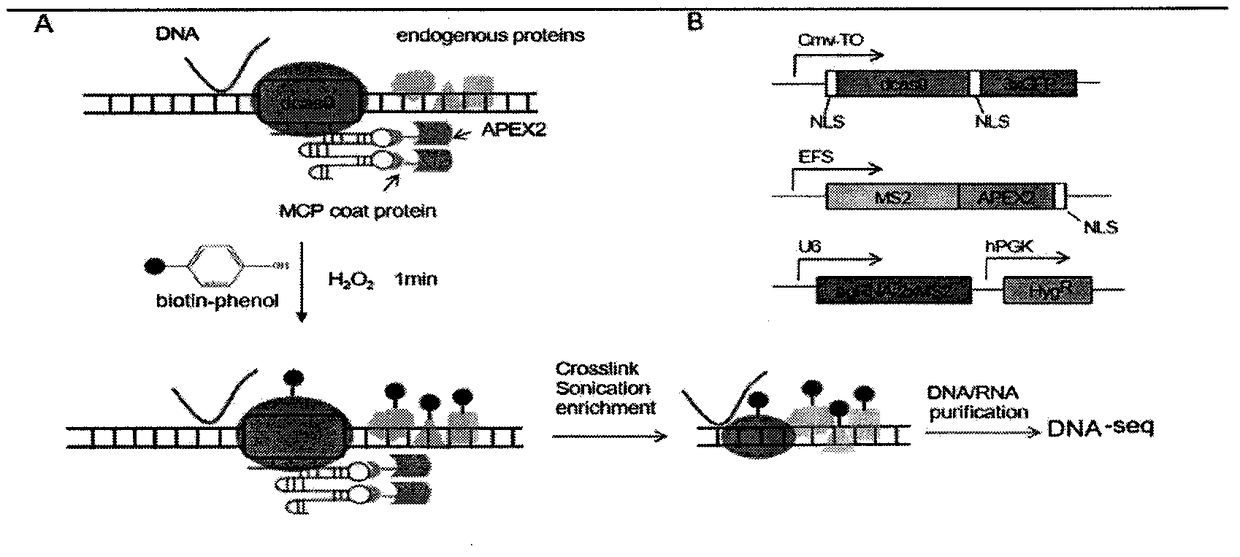
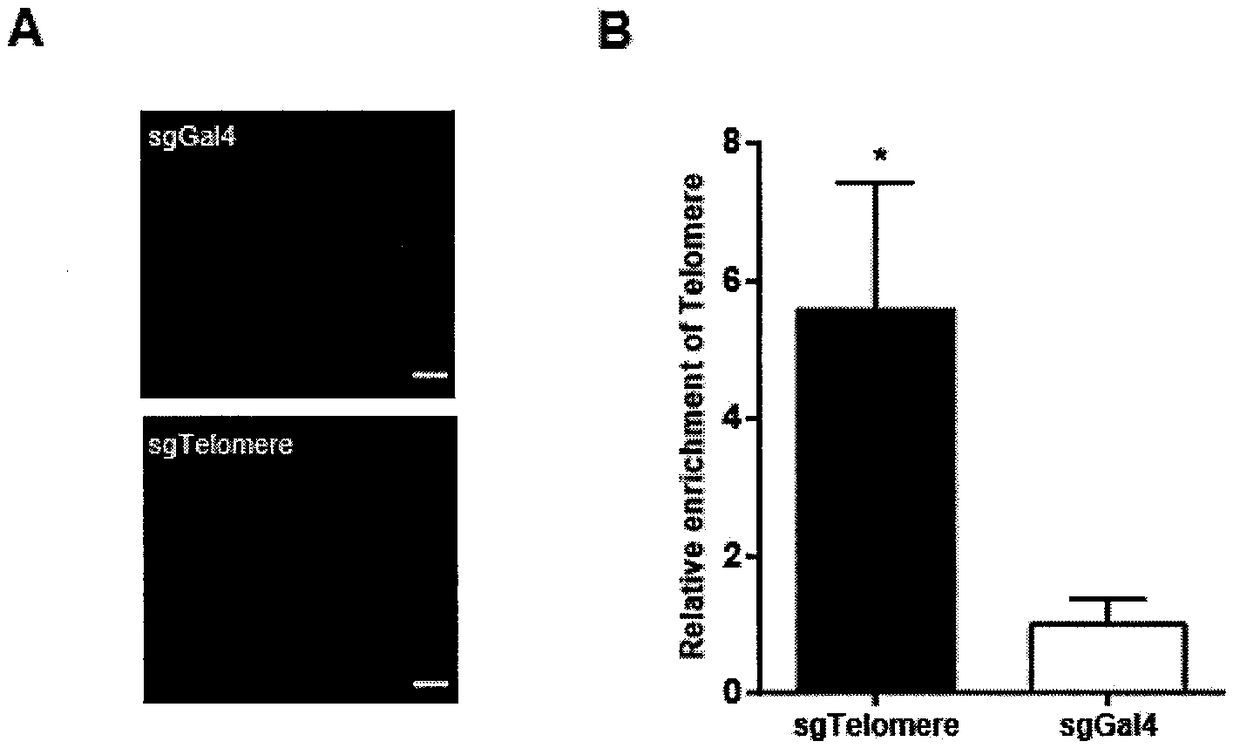
Method for identifying and analyzing specific genomic locus interaction DNAs on basis of CRISPR/cas9 and peroxidase APEX2 system
A peroxidase and specific technology, applied in the direction of recombinant DNA technology, biochemical equipment and methods, and other methods of inserting foreign genetic materials, can solve the problem of not being able to specifically provide natural chromatin or genome-wide functional analysis , Changing the natural environment of chromatin, technical difficulties and other issues, to achieve high specificity, high sensitivity and good reproducibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1, construction and transfection of plasmid expression vector
[0035] 1.1 Construction of plasmid expression vector: In order to construct MS2-APEX2_NLS fusion protein expression vector, APEX2 was amplified from pcDNA3 Connexin43-GFP-APEX2 (Addgene plasmid: 49385) plasmid by PCR method, and then it was amplified with BamHI and XhoI. Cut and cloned into the pHAGE-EFS-MCP-3XBFPnls (Addgene plasmid: 75384) vector, by connecting the annealed oligonucleotide fragment (oligos) and the BbsI restriction site to pLH-sgRNA1-2XMS2 ( Addgene plasmid: 75389) completed the construction of the sgRNA expression vector on the plasmid, and expressed dCas9 with Addgene plasmid 64107; all sgRNA sequences and are shown in Table 1;
[0036] Table 1 All sgRNA sequences
[0037] name
Sequence(5'-3')
sgRNA-Telomere
TAGGGTTAGGGTTAGGGTTA
sgRNA-Gal4
AACGACTAGTTAGGCGTGTA
sgRNA-C20-1
CTCTTAGCTGTTATGCTGTA
sgRNA-C20-2
GGATTCCCTTCCAT...
Embodiment 2
[0040] Embodiment 2, extract DNA and carry out ChIP experiment
[0041] 2.1 After 24 hours of cell transfection, when 2x107 cells were transfected with the target sgRNA or Gal4 control, the cells were treated with 500uM biotin-phenol (Iris Biotech GmbH, Germany) for 30 minutes, and then hydrogen peroxide (H 2 o 2 ) to a final concentration of 1 mM, and the cells were treated for 1 min, after which the reaction termination solution (10 mM sodium azide, 10 mM V c , 5mM Trolox) to terminate the reaction, and finally, fixed with 1% formaldehyde and incubated at room temperature for 10min. Lyse the cells with 1mL of hypotonic buffer (20mM HEPES pH7.5, 10mM potassium chloride, 1mM EDTA, 0.1mM activated sodium orthosclerate, 0.2% NP-40, 10% glycerol) for 15min, and store the cell lysate at 4°C , centrifuge at 13000g for 1min, discard the supernatant;
[0042] 2.2 Resuspend with 500ul ChIP dilution buffer (20mM Tris-HCl pH 8.0, 2mM EDTA, 150mMNaCl, 0.1% SDS, 1% TritonX-100);
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com