Salmonella bacteriophage tail receptor binding protein RBP-55 and application thereof in detection of salmonella
A technology of RBP-55, receptor binding, applied in the direction of phage, virus/phage, application, etc., to achieve the effect of expanding the scope of application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1 Extraction of Salmonella phage STP55 genome
[0066] Pick a single colony of Salmonella (ATCC 14028) from the solid medium, shake and culture at 37°C for 3h to OD 600nm 0.8-1.0, add 5mL of phage STP55 suspension, shake and culture at 37°C for 4h-6h until the culture medium changes from turbid to clear, take 1mL of high-titer phage suspension, add deoxyribonuclease and ribonuclease RNase in sequence A, incubate at 37°C for 40 min; add 20 μL 2mol / L ZnCl 2 , incubate at 37°C for 7 minutes; after centrifugation, dissolve the pellet in 500 μL TES buffer, and place in a water bath at 65°C for 15 minutes; digest with proteinase k (20 mg / mL) and cool, add 60 μL pre-cooled 3 mol / L potassium acetate, and place on ice for 15 minutes After centrifugation, the supernatant was extracted with phenol / chloroform / isoamyl alcohol (25:24:1) to extract DNA, washed with 70% ethanol and then dissolved in TE buffer to obtain phage genome samples. -Generation Sequencing, NGS) to mea...
Embodiment 2
[0068] The construction of embodiment 2 recombinant plasmids
[0069] Add the predicted orf 55 with a His tag at the N-terminus, design primers to introduce Xho I and NdeI restriction endonuclease sequences on both sides of orf 55, and combine the fragment with the empty vector pET28b (+) with Xho I and Nde I 37 Double enzyme digestion at ℃ overnight, the digestion product was recovered after verification on agarose gel electrophoresis, orf 55 was mixed with the double digestion product of pET28b(+), ligated with T4 DNA ligase overnight at 16 °C, and the ligation product Transformed into Escherichia coli T7E competent cells by heat shock method. Pick a single colony in LB medium, culture it overnight and send it for sequencing, and determine the correct sequence as a positive transformant.
Embodiment 3
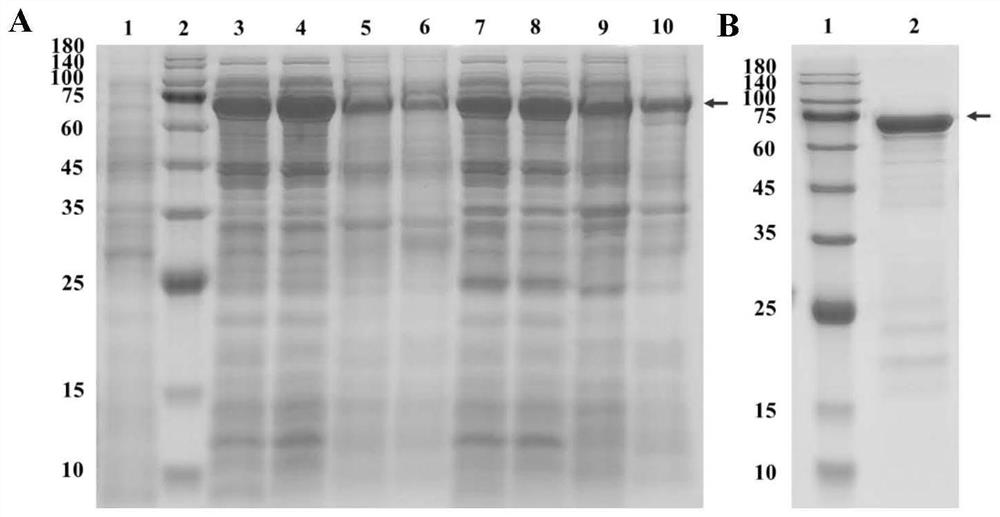
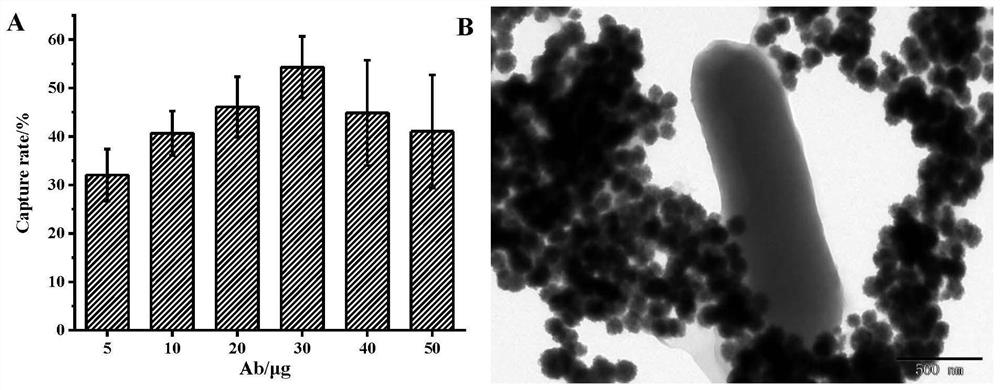
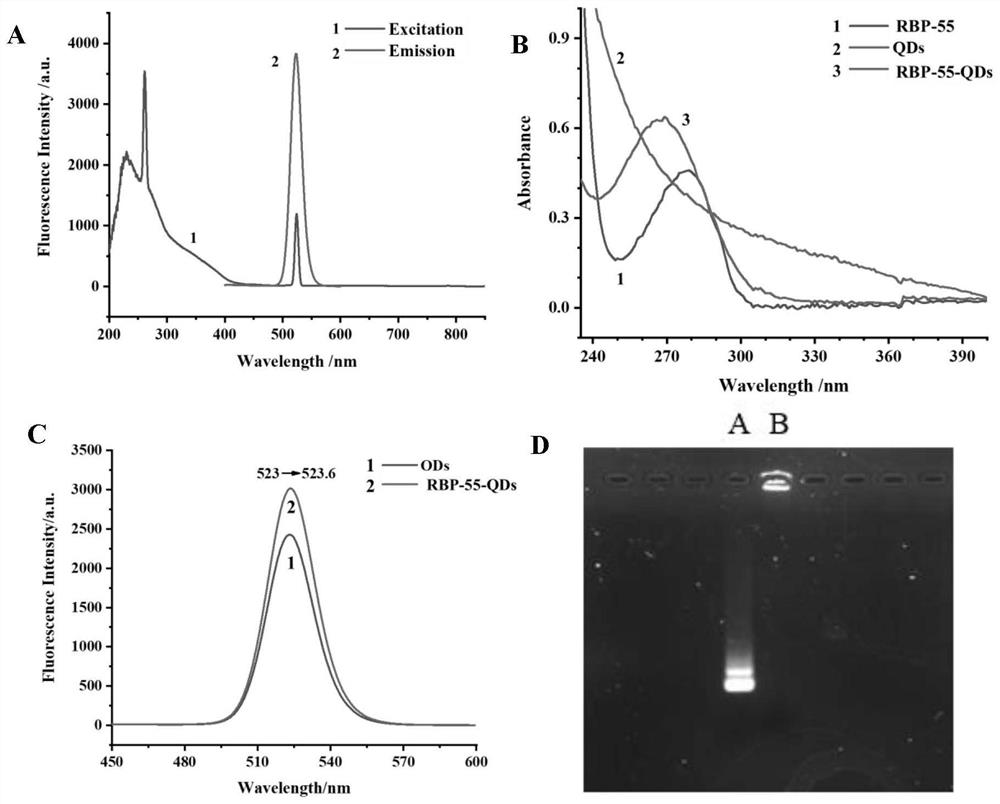
[0070] Example 3 Induced Expression and Purification of Recombinant Phage Receptor Binding Protein
[0071] The transformed competent cells were evenly spread on LB agar medium containing kanamycin (50 μg / mL), and cultured overnight at 37°C. Pick a single colony containing the recombinant plasmid and inoculate it in 5 mL LB medium containing kanamycin (50 μg / mL), and culture overnight at 37°C. Transfer 200μL to 20mL LB medium containing resistance, culture at 37℃ to OD 600nm =0.6-0.8, add IPTG (final concentration 1mmol / L) and culture at 37°C for 4h. The cells were resuspended in PBS, the cells were disrupted by ultrasonication, and samples were taken for SDS-PAGE to verify the expression status. Equilibrate the nickel column to room temperature, equilibrate with the washing solution, pass the collected supernatant sample through the column, wash the nickel column with a buffer containing 20mM imidazole, elute the target protein with an eluent of 250mM imidazole, and dialyze...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| fluorescence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com