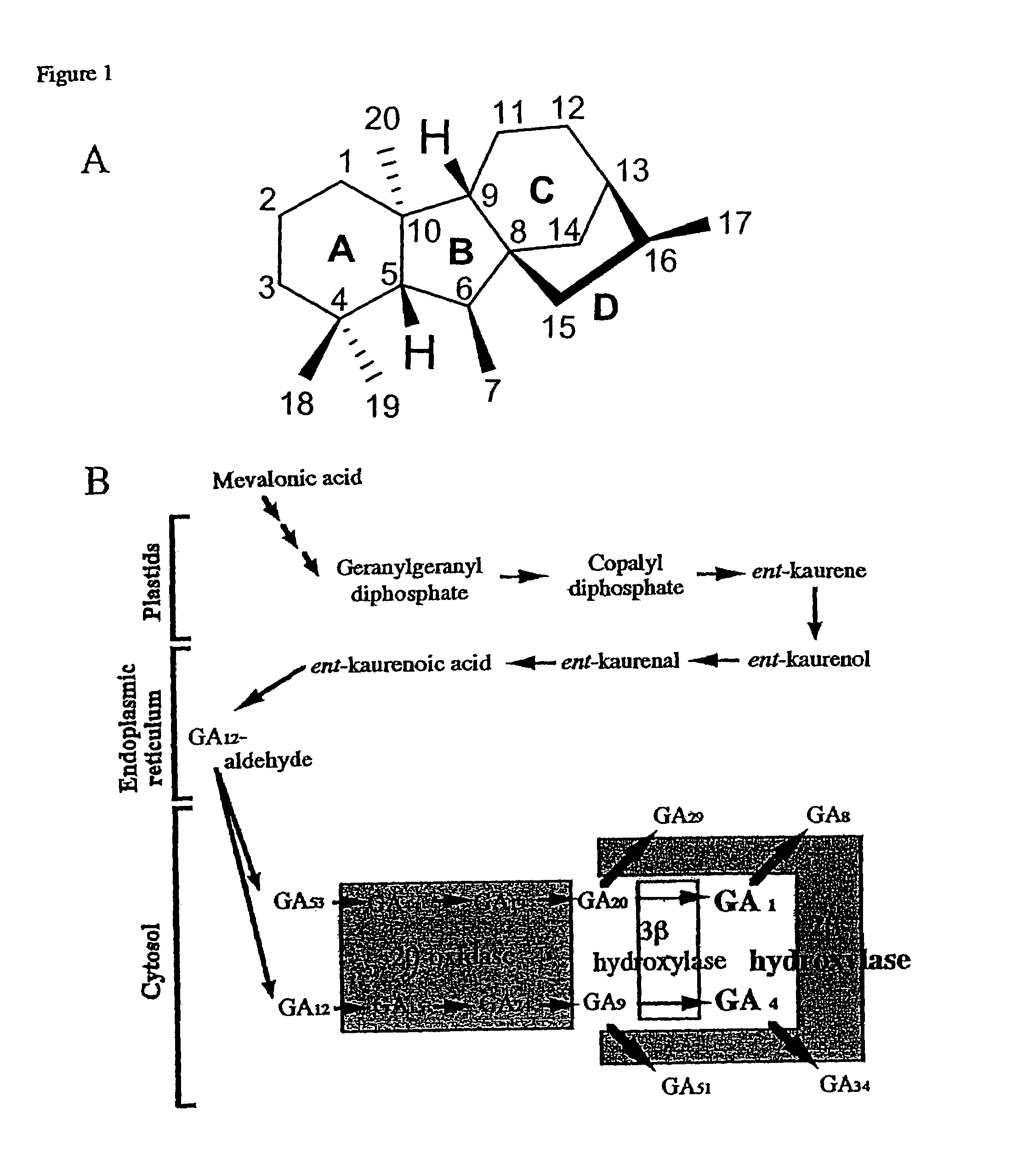
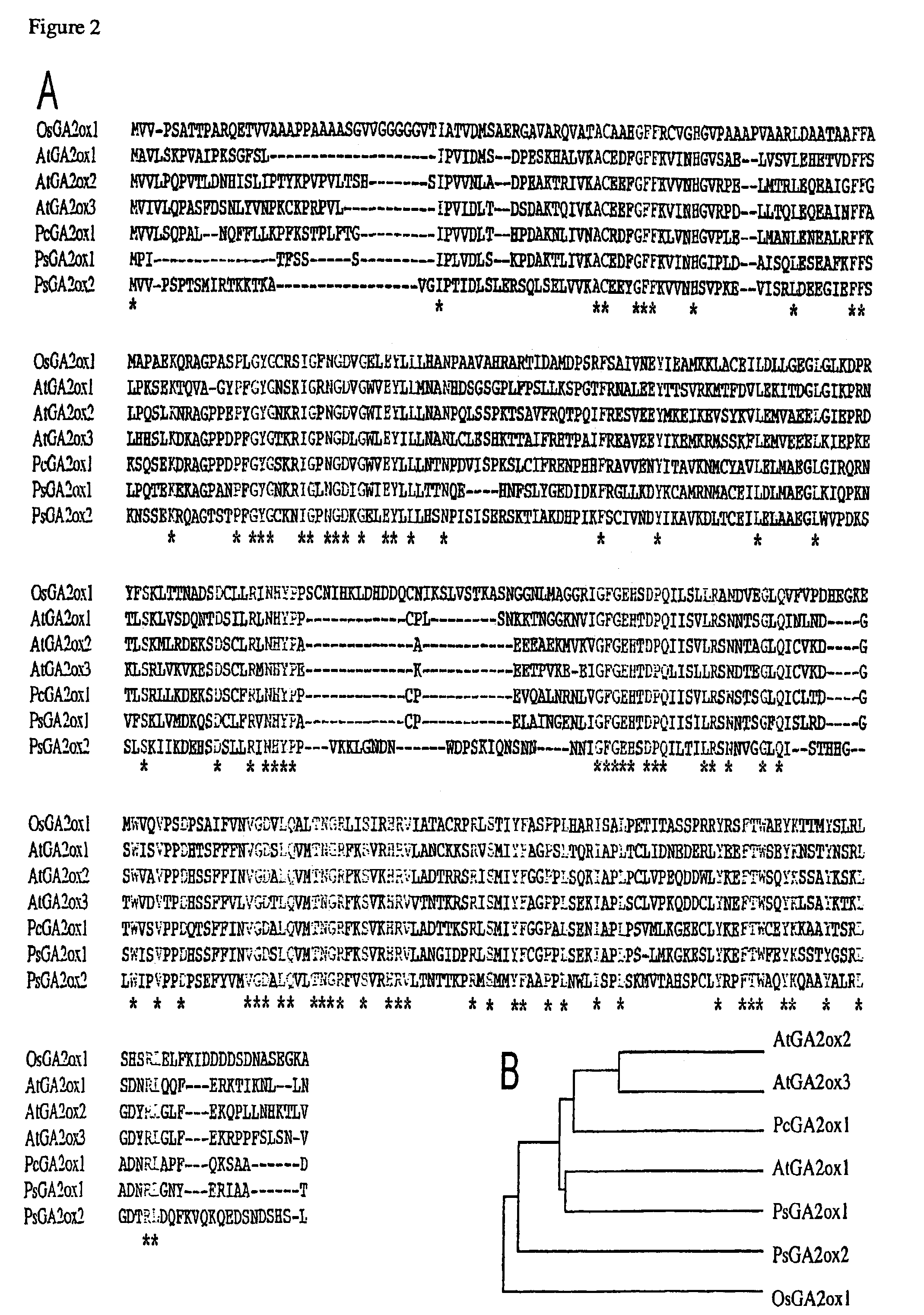
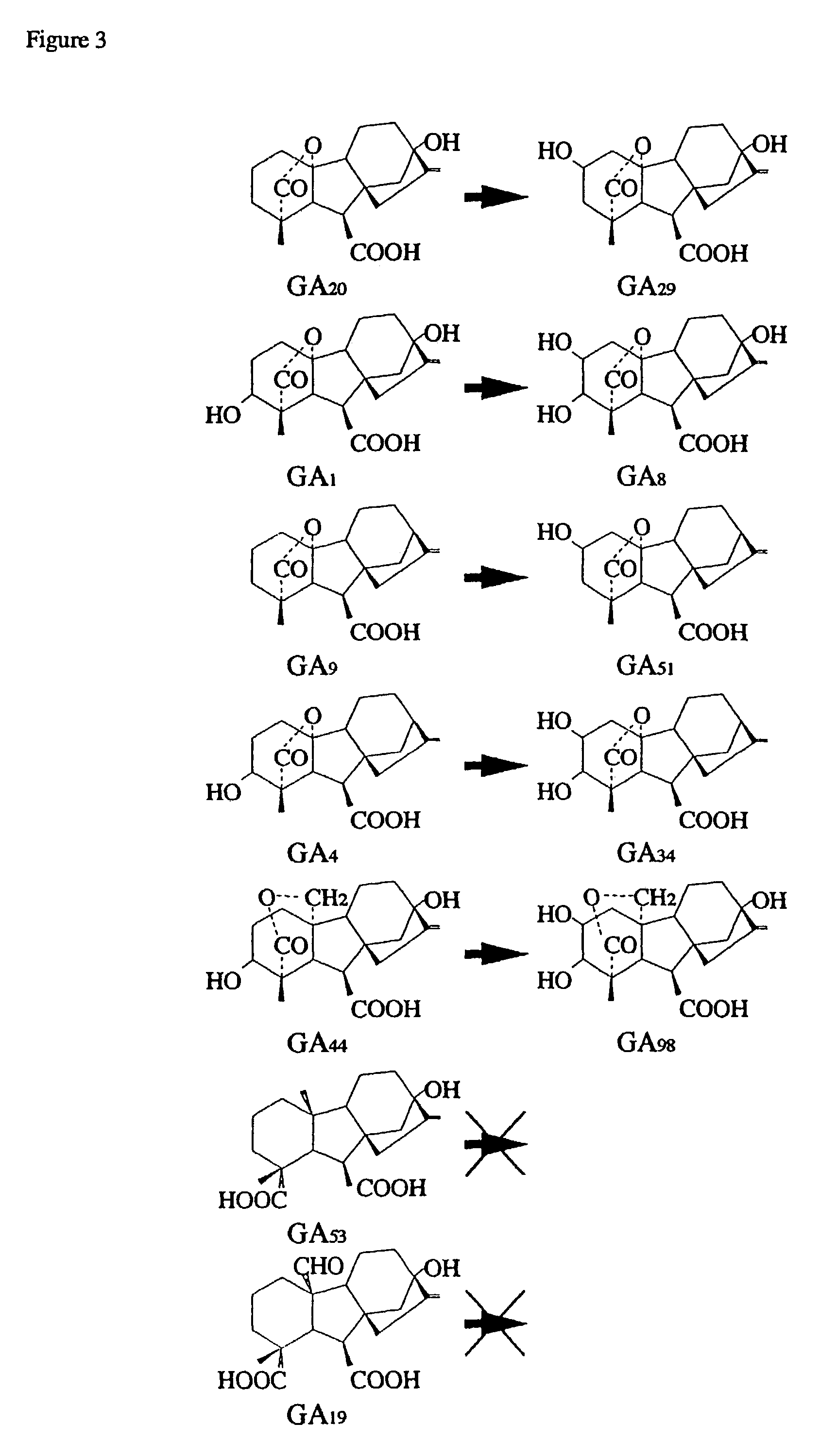
Gibberellin 2β-hydroxylase genes of rice and uses thereof
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Isolation of GA 2β-hydroxylase Gene from Rice
[0078]To amplify genomic DNA from rice (Oryza sativa L.) japonica cv. Nihon-bare, two degenerate oligonucleotide primers were designed from the conserved region of putative Arabidopsis GA 2β-hydroxylase gene (AtGA2ox3) (cDNA corresponding to DDBJ accession number C72618), Marah macrocarpa mRNA for dioxygenase (accession number Y09113; MacMillan, J. et al. (1997). Plant Physiol., 113, 1369–1377), rice GA 20-oxidase gene (accession number U50333; Toyomasu, T. et al. (1997). Physiol. Plant., 99, 111–118), rice GA 3β-hydroxylase genes, and other 2-oxoglutarate-dependent dioxygenase genes (forward primer, 5′-GGNTTYGGNGARCAYWCNGAYCC-3′ / SEQ ID NO: 3; and reverse primer, 5′-GGISHISCRAARTADATIRTISWIA-3′ / SEQ ID NO: 4). PCR was performed using rice genomic DNA as a template. The amplified fragments (about 80 bp) were cloned into pCR II (Invitrogen, Carlsbad, Calif.) and their sequences were confirmed. One of the 64 independent clones contained a nov...
example 2
Function of Recombinant GA 2β-Hydroxylases
[0082]The full-length cDNA of rice GA 2β-hydroxylase was inserted in the sense orientation as a translational fusion into the pMAL-c2 expression vector (New England Biolabs, Beverly, Mass.). The resulting construct, pMAL-OsGA2ox1, was expressed in Escherichia coli strain JM109. Bacterial cells were grown overnight at 30° C. in 2×YT medium containing 0.2% [w / v] glucose and 100 mg / L ampicillin. After overnight growth, cultures were diluted 500-fold with the fresh medium and incubated with shaking at 30° C. When growth reached an OD600 of 0.7, IPTG was added to a final concentration of 1 mM, and culturing was resumed at 17° C. for a period of further 24 hr. These bacterial cells were harvested, washed with washing buffer (50 mM Tris, pH 8.0, 10% [w / v] glycerol, 2 mM DTT), resuspended in the washing buffer containing 1 g / L lysozyme, and kept on ice for 30 min.
[0083]The lysate thus obtained was sonicated and centrifuged. Its supernatant was subje...
example 3
Expression of GA 2β-Hydroxylase Gene in Rice
(1) RNA Gel Blot Analysis
[0084]Total RNAs from rice were separately prepared from various tissues (vegetative shoot apices, young leaves, stems, leaf blades, leaf sheath, root, inflorescence shoot apices, glumes, and rachis) for RNA gel blot analysis. Ten μg of each RNA preparation was electrophoresed on a 1.2% agarose gel, transferred onto Hybond N+ membrane (Amersham, Buckinghamshire, England), and then hybridized with the HindIII-EcoRV fragment (the 230 bp fragment of OsGA2ox1 cDNA) as a probe. Hybridization was performed in 5×SSC, 5×Denhardt's solution, 0.5% [w / v] SDS, and 20 mg / L salmon sperm DNA at 65° C. for 14 hr. The filter was washed in 2×SSC, 0.1% [w / v] SDS at 65° C. and then further washed in 0.2×SSC, 0.1% [w / v] SDS at 65° C.
[0085]A single strong band was detected in RNA from all organs examined (FIG. 4). The size of the band was ca. 1.6 kb that was almost the same size as the cDNA clone.
[0086]To more p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com