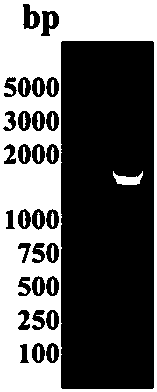Endo-cellulase coding gene as well as preparation and application thereof
A technology of endo-cellulase and cellulose, which is applied in application, genetic engineering, plant genetic improvement, etc., can solve the problems of less hemicellulose substrates, weak konjac polysaccharide degradation activity, and unsuitable for large-scale application, achieving The effect of efficient degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Embodiment 1 endocellulase full-length gene cloning
[0056] Genomic DNA of Paenibacillus polymyxa was extracted according to the operation steps of Genomic DNA Purification Kit (Thermo, LOT 00105781). After multiple sequence alignment analysis of endocellulase gene sequences in The National Center for Biotechnology Information (NCBI) database, degenerate primers were designed Ppcell-F: 5'-CGGACGCATATGGAATCARCTMMCTTGGTCAC-3'; Ppcell-R: 5'-GACGAGCTCGAGCTCCGCTTYATTYTTGGABRAAGTA -3', using the extracted Paenibacillus polymyxa genomic DNA as a template to amplify the gene sequence encoding the endocellulase mature protein (excluding the signal peptide gene). The PCR reaction conditions were as follows: 94°C for 3min, 1 cycle; 94°C for 30s, 55°C for 30s, 72°C for 1min30s, 30 cycles; 72°C for 5min, 1 cycle. PCR products were analyzed by agarose gel electrophoresis (see figure 1 ), the target gene was recovered by cutting the gel, connected to the prokaryotic expression vect...
Embodiment 2
[0057] Embodiment 2 endocellulase gene sequence analysis
[0058] The sequencing results were analyzed using Basic Local Alignment Search Tool (BLAST) in the GenBank database, DNAMAN software was used for multiple sequence alignment, and Vector NTI was used to analyze sequence information.
[0059] The coding region of the obtained endocellulase gene (named Ppcell) is 1662 bp long, and its nucleotide sequence is shown in SEQ ID NO 1. Ppcell encodes 553 amino acids and a stop codon, its amino acid sequence is shown in SEQ ID NO 2, the theoretical molecular weight of the protein is 61.45kDa, and the predicted isoelectric point is 6.55. The amino acid encoded by Ppcell contains a GH1 family domain and a carbohydrate binding module CBM (Carbohydrate Binding Module) X2 domain, and its GH1 domain overlaps with the cellulase domain of the GH6 family, thus indicating that Ppcell is a Dual-domain functional enzyme.
Embodiment 3
[0060] Recombinant expression and purification of embodiment 3 Ppcell gene in Escherichia coli
[0061] In order to facilitate the recombinant expression of the gene, NdeI and XhoI restriction sites were respectively introduced into the designed upstream and downstream primers. The PCR cleaning product Ppcell and the expression vector pET21a were double-digested with NdeI and XhoI respectively. 4 DNA ligase connection (ligation system: (5μLT 4 DNA Ligase 0.5μL, 10×T 4DNA LigaseBuffer 0.5 μL, pET21a 2 μL, PCR product 2 μL), ligation conditions: overnight at room temperature. ). Take 5 μL of the ligation product to transform E.coli TOP10 competent cells, spread on solid Luria-Bertani medium containing 100 μg / mL ampicillin, and culture at 37°C for 12-16h. Pick a single clone, use degenerate primers for colony PCR verification, insert the correctly amplified single clone into liquid Luria-Bertani medium containing 100 μg / mL ampicillin, and extract the plasmid; use endonuclease...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com