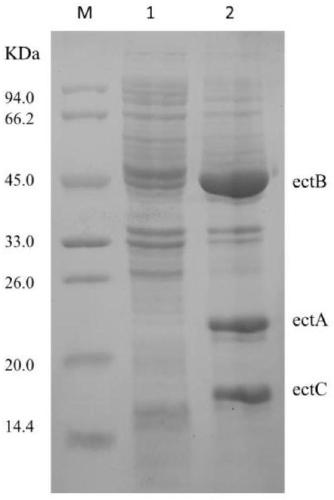
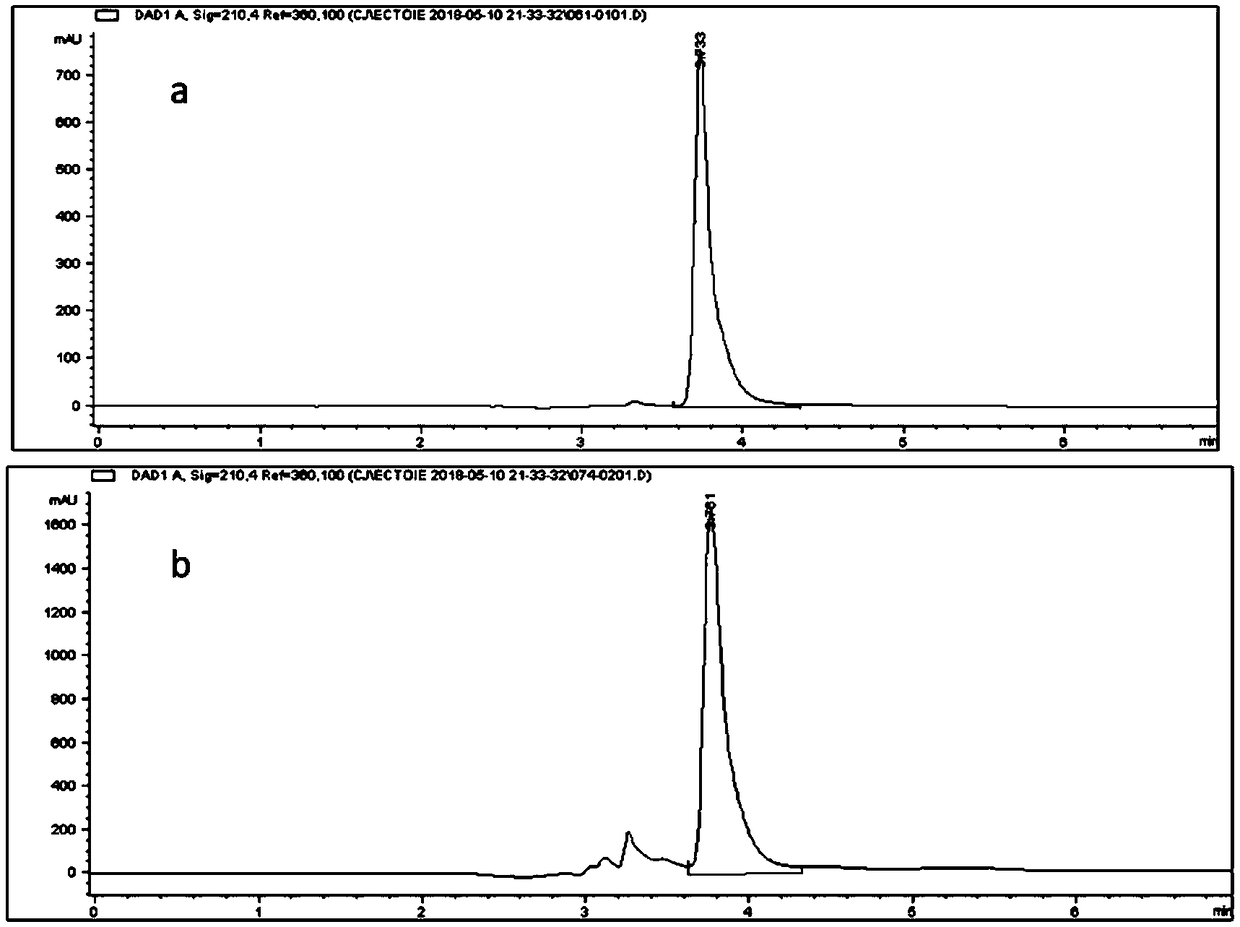
Recombinant escherichia coli and application to synthesis of tetrahydropyrimidine
A technology for recombining Escherichia coli and tetrahydropyrimidine is applied in the fields of genetic engineering and biology, and can solve the problems of difficulty in advancing the research progress of tetrahydropyrimidine, low yield of tetrahydropyrimidine and tetrahydropyrimidine, and high cost of chemical synthesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Construction of Escherichia coli ectoine metabolic pathway
[0026] 1. Knockout of lysA gene
[0027] Gene manipulation method using Red homologous recombination
[0028] According to the lysA gene sequence and pKD3 plasmid sequence of E. coli MG1655, primers lysA-1 and lysA-2 were designed to amplify the resistant fragment using pKD3 as a template. The program of PCR amplification: 30 cycles, denaturation at 98°C for 30 sec, annealing at 55°C for 15 sec, extension at 72°C for 30 sec.
[0029] Table 1 PCR amplification system
[0030]
[0031] Table 2 Primer Nucleic Acid Sequence
[0032]
[0033] Electrotransform the amplified resistant fragment (see SEQ ID NO.2 for the nucleotide sequence) into E.coli MG1655 containing the PkD46 plasmid, and spread it on a plate containing chloramphenicol resistance (30 μg / mL) , Colony PCR identification and screening of positive transformants, that is, the lysA knockout strain E.coli MG1655-1 was obtained.
[00...
Embodiment 2
[0042] Embodiment 2: Engineering bacterium E.coli Hect fermentation preparation ectoine
[0043] 1. Fermentation of engineering bacteria E.coli Hect
[0044] The engineered bacteria E.coli Hect, E.coliMG1655-1-pTrc99a, and E.coli MG1655 were inoculated in the seed medium for shaker culture respectively. The culture conditions: temperature 37°C, rotation speed 200rpm, culture time 12h to logarithmic growth period to obtain seed solution. Seed medium composition: tryptone 10g / L, yeast extract 5g / L, sodium chloride 10g / L, solvent is deionized water, pH7.0.
[0045] The seed solution was added to the fermentation medium with an inoculation amount of 10% volume concentration, and the culture temperature was 37 ° C, 200 rpm shaking culture until the OD600 was 0.6, 50 mg / mL inducer IPTG was added, and the culture temperature was 30 ° C, 200 rpm shaking culture for 12 hours, Obtain a fermented broth. After the fermentation, the fermentation broth was centrifuged at 5000 rpm at 4° C...
Embodiment 3
[0050] Embodiment 3: Optimization of the whole cell biocatalysis system
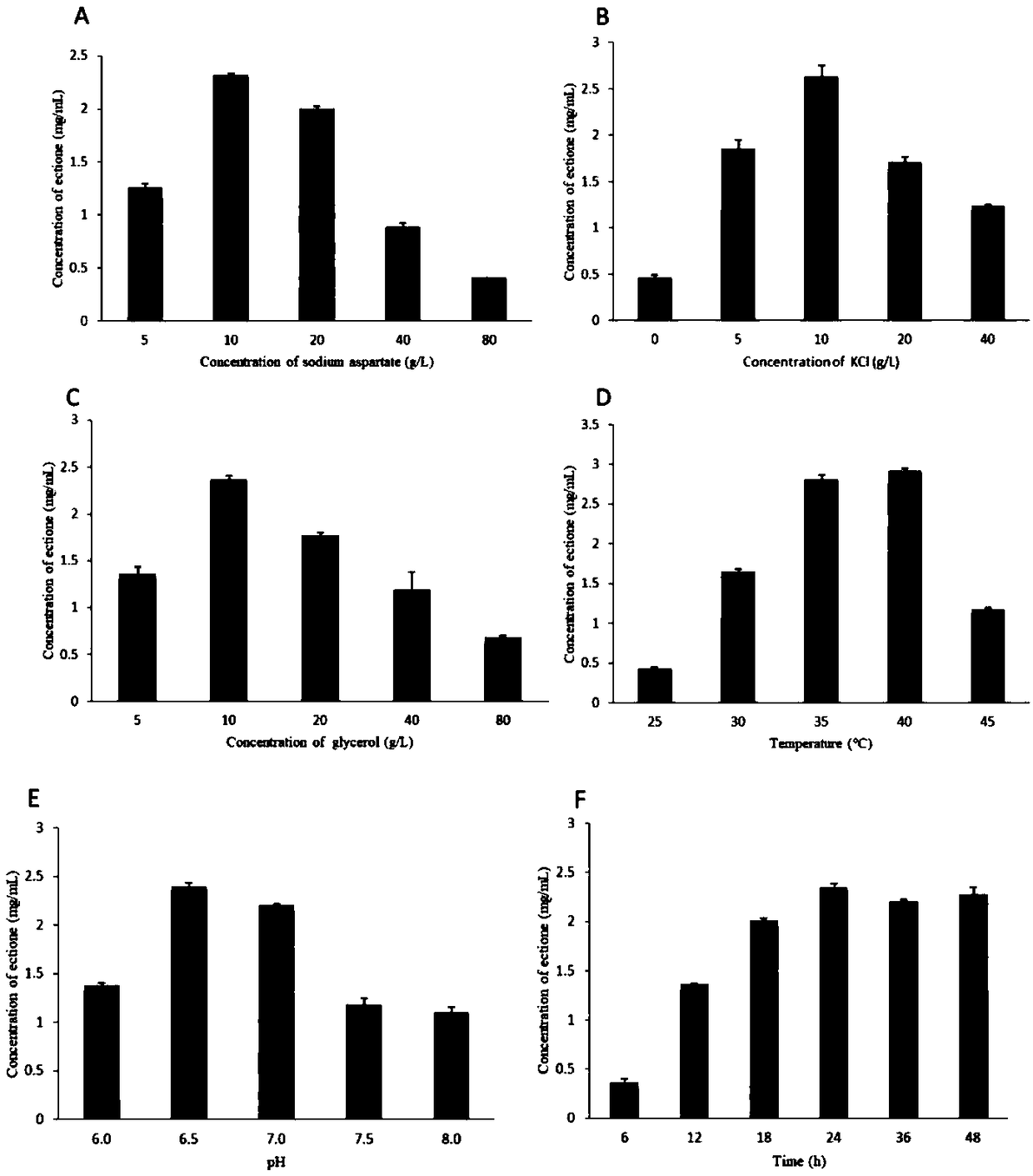
[0051] Centrifuge to collect the bacteria after fermentation 12 in Step 1 of Example 1, and resuspend the bacteria to OD with PBS buffer 600 =5, on the basis of Example 2, investigate the influence of different conversion liquid compositions and reaction temperature and time on the preparation of ectoine respectively, according to Example 2 conversion liquid (L-sodium aspartate 10g / L, glycerol 10g / L, Potassium Chloride 10g / L, solvent is PBS buffer solution, pH6.5), investigates the influence of one element at a time, all the other element concentrations are constant.
[0052] Sodium L-aspartate (5, 10, 20, 40, 80g / L), potassium chloride (0, 5, 10, 20, 40g / L), glycerin (5, 10, 20, 40, 80g / L) L), pH value (6.0, 6.5, 7.0, 7.5, 8.0), reaction temperature (25, 30, 35, 40, 45°C), reaction time (6, 12, 18, 24, 36, 48h).
[0053] The result is as image 3 As shown, the best whole-cell biocatalytic system is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com