A kind of α-amylase, encoding gene, carrier, host and application thereof
A technology encoding gene and amylase, applied in the fields of bioengineering and enzyme engineering, can solve problems such as the inability to faithfully reflect the function of functional enzyme genes, and the inability to effectively mine enzyme genes by culture methods.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: Acquisition of α-amylase encoding gene
[0030]Samples were taken at the high temperature stage (62°C) of Luzhou-flavor Daqu, and total RNA was extracted. The specific method is as follows: grind 1 g of Luzhou-flavor Daqu sample into a fine powder in a pre-cooled mortar. The sample was mixed with 4 mL of borate buffer (200 mM sodium borate, 30 mM ethylene glycol tetraacetic acid (EGTA), 1% (w / v) sodium dodecyl sulfate (SDS), 4% (w / v) v) Polyvinylpyrrolidone (PVP), 0.5% (v / v) Nonidet-40 (NP-40), 10 mM β-mercaptoethanol and 0.03% (v / v) RNase inhibitor) and 280 μL proteinase K (20 mg / mL) Mix and keep at room temperature for 2 minutes. RNA from the resulting crude lysate was then centrifuged, precipitated with 70% ethanol, and washed according to the instructions of the RNeasy Midi Kit (Qiagen, Valencia, CA). In addition, the RNA mixture was treated with DNase I (Fermentas, USA) according to the instructions. Use 3 μL of total RNA (approximately 1 μg) as a te...
Embodiment 2
[0031] Example 2: α-amylase bioinformatics analysis
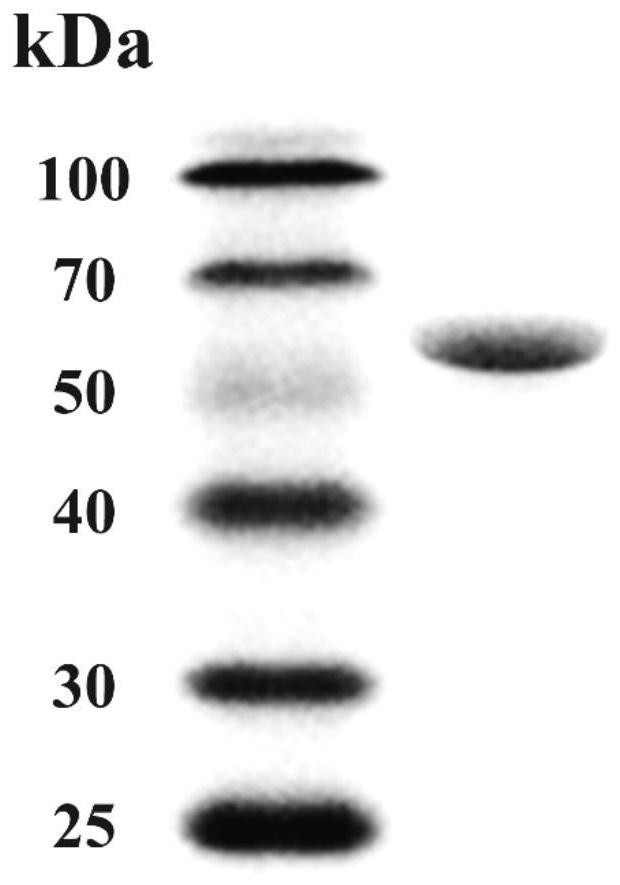
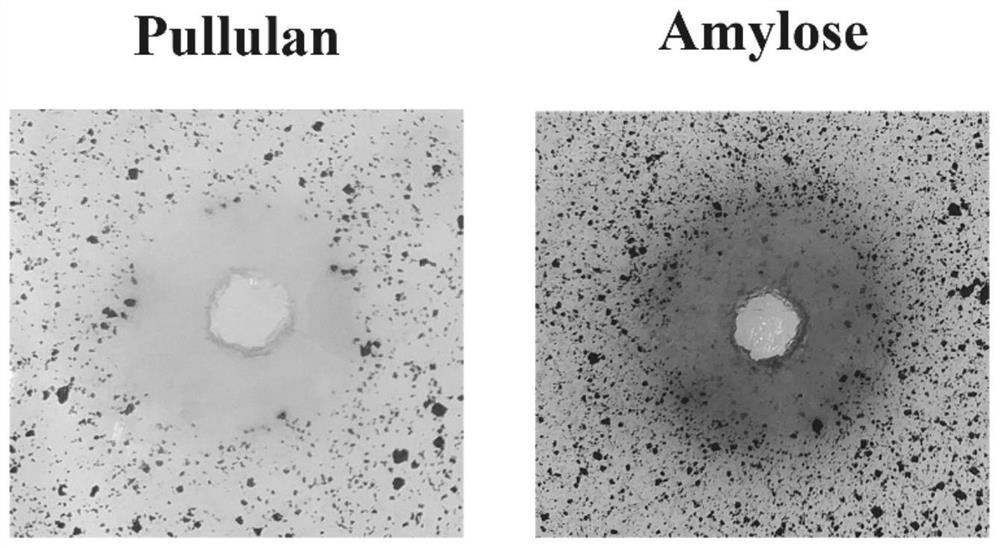
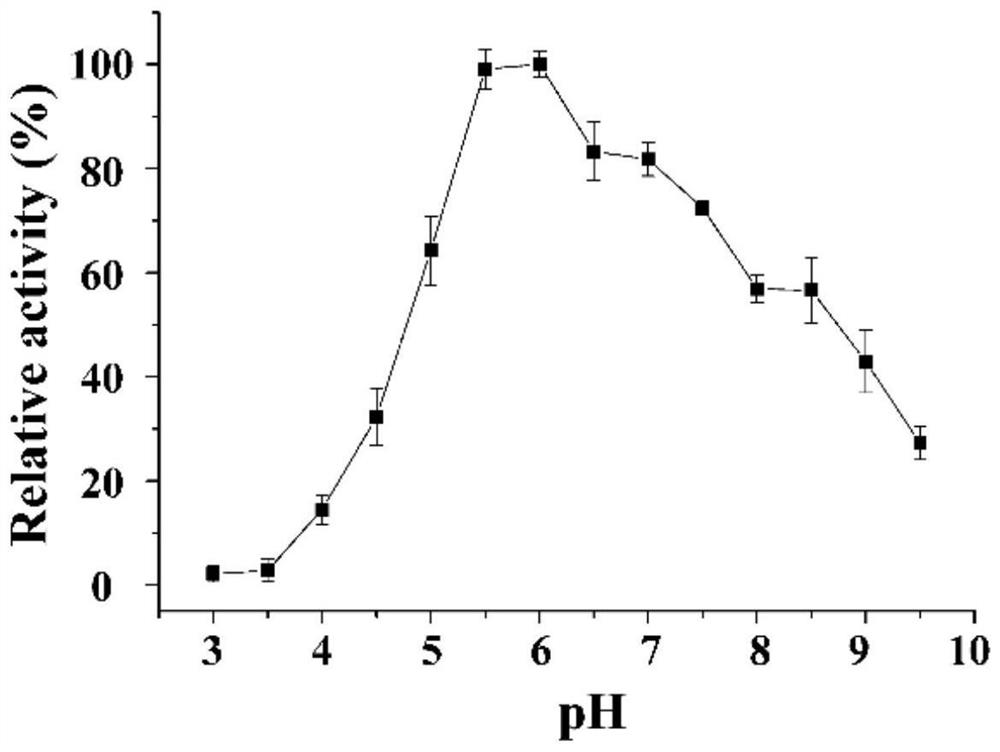
[0032] The protein expressed by the NFAmy13B gene obtained in Example 1 is named α-amylase NFAmy13B, which contains 534 amino acids, its amino acid sequence is shown in SEQ ID NO.2, and the predicted protein size is 61.2KD. The nucleotide sequence of NFAmy13B gene was predicted according to SignalP-5.0Server (http: / / www.cbs.dtu.dk / services / SignalP / ), which showed that it had no signal peptide. According to the gene bank sequence comparison, the enzyme with the highest amino acid sequence similarity to the α-amylase NFAmy13B is a putative α-amylase derived from Byssochlamys spectabilis (GenBank number: XP_028489551.1), with a similarity of 77.6%. Amylase was derived from the analysis of genome information, and the enzymatic properties were not analyzed. The second is the intracellular fungal amylase AmyD (GenBank number: XP_001389762.2) derived from Aspergillus niger, with a similarity of 64.4%. Phylogenetic tree analysis ...
Embodiment 3
[0033] Example 3: Expression of α-amylase gene
[0034] NFAmy13B was amplified by forward primer NFABf (5'-CACCATGAAGTCCCTCCTCTGCTG-3') and reverse primer NFABr (5'-CTAGTGCTTGTAGATATCCGAGTC-3') using the cDNA containing open reading frame 22984 in the Luzhou-flavor Daqu cDNA library as a template Gene. The specific PCR reaction system is: 47 μL of Mix (green) (TsingKe, Beijing), 1 μL of 10 μM forward and reverse primers, and 1 μL of DNA template. PCR amplification conditions were: pre-denaturation at 98°C for 2 min, denaturation at 98°C for 10 s, annealing at 50°C for 15 s, extension at 72°C for 25 s, skip to step 2 and cycle 30 times, and keep at 72°C for 5 min. The amplified product detected by agarose gel electrophoresis was about 1.6kb, which was consistent with the expected NFAmy13B gene containing 1605 bases. 50-100 ng of the amplified product was ligated with pDE2 plasmid (TsingKe, Beijing) at 22-30° C. for 1-5 min to construct a recombinant expression vector.
[003...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com