Nitrogen-Regulated Sugar Sensing Gene and Protein and Modulation Thereof
a sugar sensing gene and nitrogen-regulated technology, applied in the direction of sugar derivatives, plant cells, angiosperms/flowering plants, etc., can solve the problems of increasing the cost of this input to the farmer, increasing the cost of this input to the environment, and reducing the effect of resistance to herbicides, enhancing or reducing the requirement for light, water, nitrogen or trace elements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
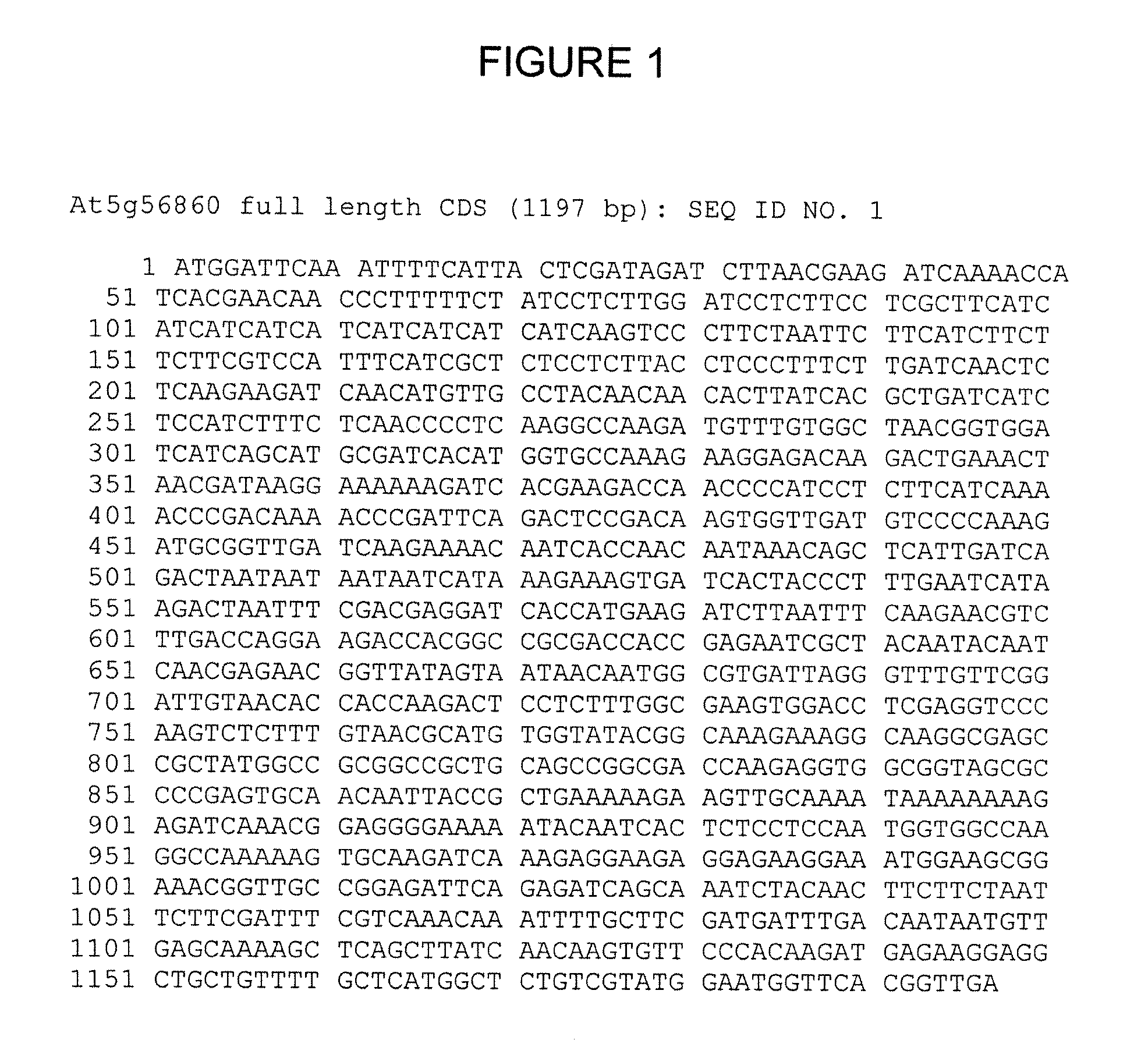
Cloning and Sequence of At5g56860
[0420] Gene predictions were derived from the sequence databases, and used to design oligonucleotide primers for PCR amplification of either full-length (inclusive of the predicted initiation and stop codons, for transgenic gene overexpression) or partial (for transgenic gene knockout) cDNA clones from rice first strand cDNA. In some instances, these PCR primers included additional 5′ sequences for Gateway™ recombination-based cloning (Invitrogen). PCR amplification was carried out using the HF Advantage II (Clonetech) or EXPAND (Roche) PCR kits according to the manufacturer's instructions. PCR products were cloned into pCR2.1-TOPO or pDONR201 according to the manufacturer's instructions (Invitrogen).
[0421] DNAs from 4-8 independent clones were miniprepped following the manufacturer's instructions. DNA was subjected to sequencing analysis using the BigDye™ Terminator Kit according to manufacturer's instructions (ABI). Sequencing made use of primer...
example 2
[0441] The full length At5g56860 cDNA (GNC) was amplified from Arabidopsis leaf cDNA using the primers 5′-GCTCTAGATTTCTCTCTCTCTTTGTGTCTTCATTTG-3′ (SEQ ID NO:4) and 5′-gcgagctctcgggtgactaatgttcgttcc-3′ (SEQ ID NO:5). The resulting ˜1500 bp fragment was verified by sequencing and then digested by XbaI and SacI and cloned into the XbaI-SacI-digested expression vector pROK2 containing the cauliflower mosaic virus (CaMV) 35S promoter driving the GNC expression in a constitutive, high-level fashion (FIG. 4). The p35S-GNC binary vector was transformed into Agrobacterium tumefaciens strain EHA105 and the resulting Agrobacterium strain was used to transform the wild type plants (Col-0) and the transformants were selected on kanamycin resistance. Plants with a single insertion of the transgene were selected for self pollination to generate T3 homozygous lines for further analysis. Over-expression of GNC in the transgenics was confirmed by quantitative RT-PCR. Wild type plants and GNC over-exp...
example 3
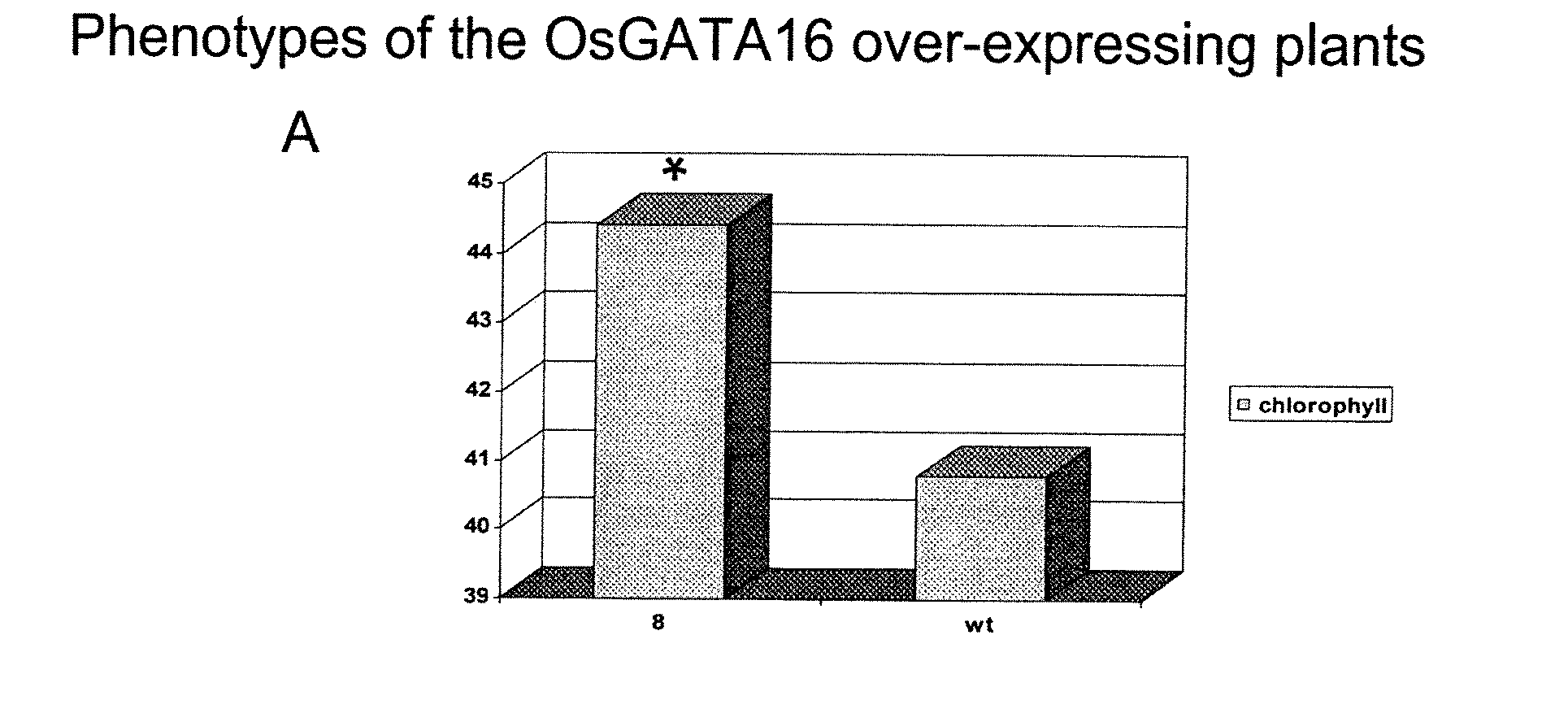
[0442] As mentioned in Example 1, the inventors identified the Arabidopsis GATA transcription factor gene GNC (At5g56860) important in chlorophyll synthesis and sugar sensitivity. The At4g26150 gene is a GNC paralog in the phylogenetic tree of the 30 Arabidopsis GATA transcription factor genes (Reyes, J. C., Muro-Pastor, M. I. & Florencio, F. J. (2004) Plant Physiol. 134, 1718-1732) and was found to have overlapping function with GNC (unpublished results). In the rice (Oryza sativa) genome, there are 28 GATA transcription factor genes, with one pair of genes (OsGATA16 and OsGATA11) sharing similarity with the two Arabidopsis GATA genes (Reyes, J. C., Muro-Pastor, M. I. & Florencio, F. J. (2004) Plant Physiol. 134, 1718-1732). Transgenic rice plants either over-expressing or silencing the two rice ortholog genes were generated. The phenotypes of these transgenic plants were analyzed to understand their in vivo function.
Materials and Methods
Plant Growth Conditions
PUM
| Property | Measurement | Unit |
|---|---|---|
| Nucleic acid sequence | aaaaa | aaaaa |
| Energy | aaaaa | aaaaa |
| Stress optical coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com