Heat resistance cutinase and its coding gene and expression
A technology for encoding genes and cutinase, applied in genetic engineering, plant genetic improvement, hydrolase and other directions, can solve the problems of low enzyme production by wild fungi, less research on cutinase, and no industrial production of cutinase.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
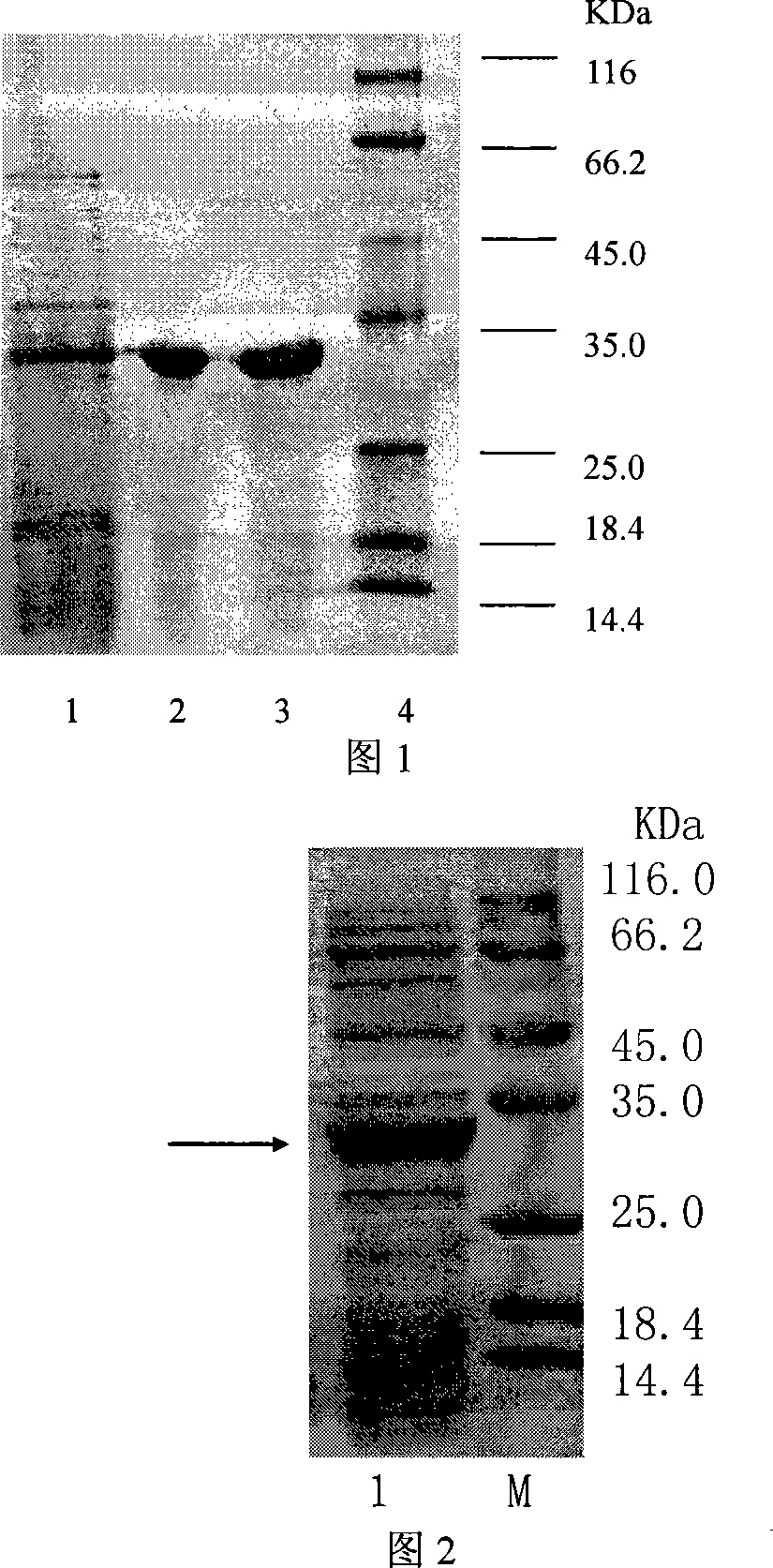
[0043] Example 1 This example illustrates the purification procedure of cutinase.
[0044] Using Thermobifida fuscaWSH03-11 as the starting strain, in the seed medium (soluble starch 20g / L, beef extract 10g / L, yeast extract 5g / L, K 2 HPO 4 2g / L, NaCl 5g / L, pH8.0) 50℃, 200rpm for 20 hours, and then inoculated into the fermentation medium (sodium acetate 7.5g / L, yeast extract 7.5g / L, peptone 5g / L, K 2 HPO 4 2g / L, NaCl 5g / L, keratin 1g / L, pH8.0), 50°C, 200rpm culture for 50h, the cutinase fermentation broth was centrifuged at 4°C, 10000rpm for 20min to remove bacteria. The supernatant was collected through an activated carbon column. Add 70% solid ammonium sulfate to the clear solution passed through the activated carbon column overnight, centrifuge at 4°C and 10,000 rpm for 20 minutes, take the precipitate and dissolve it in buffer A (20mM Tris-HCl, pH 8), add 20% ammonium sulfate, After 0.22μm membrane filtration, the loaded sample is made. After the Phenyl HP hydrophobic column i...
Embodiment 2
[0045] Example 2 This example illustrates the identification process of the cutinase gene.
[0046] The obtained pure cutinase was sequenced by peptide fingerprinting. The sequencing results showed that it was very similar to the protein Tfu_0883 encoded by Thermobifida fusca in the NCBI database. The N-terminal sequence was ANPYERGP, which was the same as the protein Tfu_0883. The protein Tfu_0883 was predicted to be triglyceride by sequence , No indication of its cutinase activity. The pure wild fungus cutinase is subjected to enzymatic hydrolysis of cutin (apple peel treated with chloroform and methanol, pectinase and cellulase). Add 300 mg of cutin to pH 8.0, 10 mL of 50 mM potassium phosphate buffer, add purified cutinase, and shake at 30°C for 18 hours. After the reaction is completed, add glacial acetic acid to terminate the reaction. The fatty acid product is extracted with chloroform and methylesterified and silanized. -MS measurement revealed that a large amount of kerat...
Embodiment 3
[0047] Example 3 This example illustrates the isolation and cloning procedure of the cutinase encoding gene.
[0048] The Thermobifida fusca strain was cultured in LB liquid medium (peptone 10g / L, yeast extract 5g / L, NaCl10g / L) for 2 days, centrifuged at 10000rpm to collect the bacteria, washed with sterile water, and collected the precipitate and suspended in 500μL Tris-EDTA (three Hydroxymethylaminomethane-ethylenediaminetetraacetic acid) buffer, add 15μL of lysozyme, incubate at 37℃ for 30min, then add 5μL of RNase, incubate at 37℃ for 30min, add 30μL of 10% SDS (sodium dodecyl sulfate) ) And 15μL of proteinase K, incubate at 37℃ for 60min, add 100μL of NaCl (5M) and 80μL CTAB (hexadecyltrimethylammonium bromide), incubate at 65℃ for 20min, use 700μL of phenol: chloroform: isoamyl alcohol (25:24:1) extraction, centrifugation at 10000rpm, the supernatant was extracted with 700μL of chloroform:isoamyl alcohol (24:1), centrifuged at 10000rpm, the supernatant was mixed with 1400μL ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com