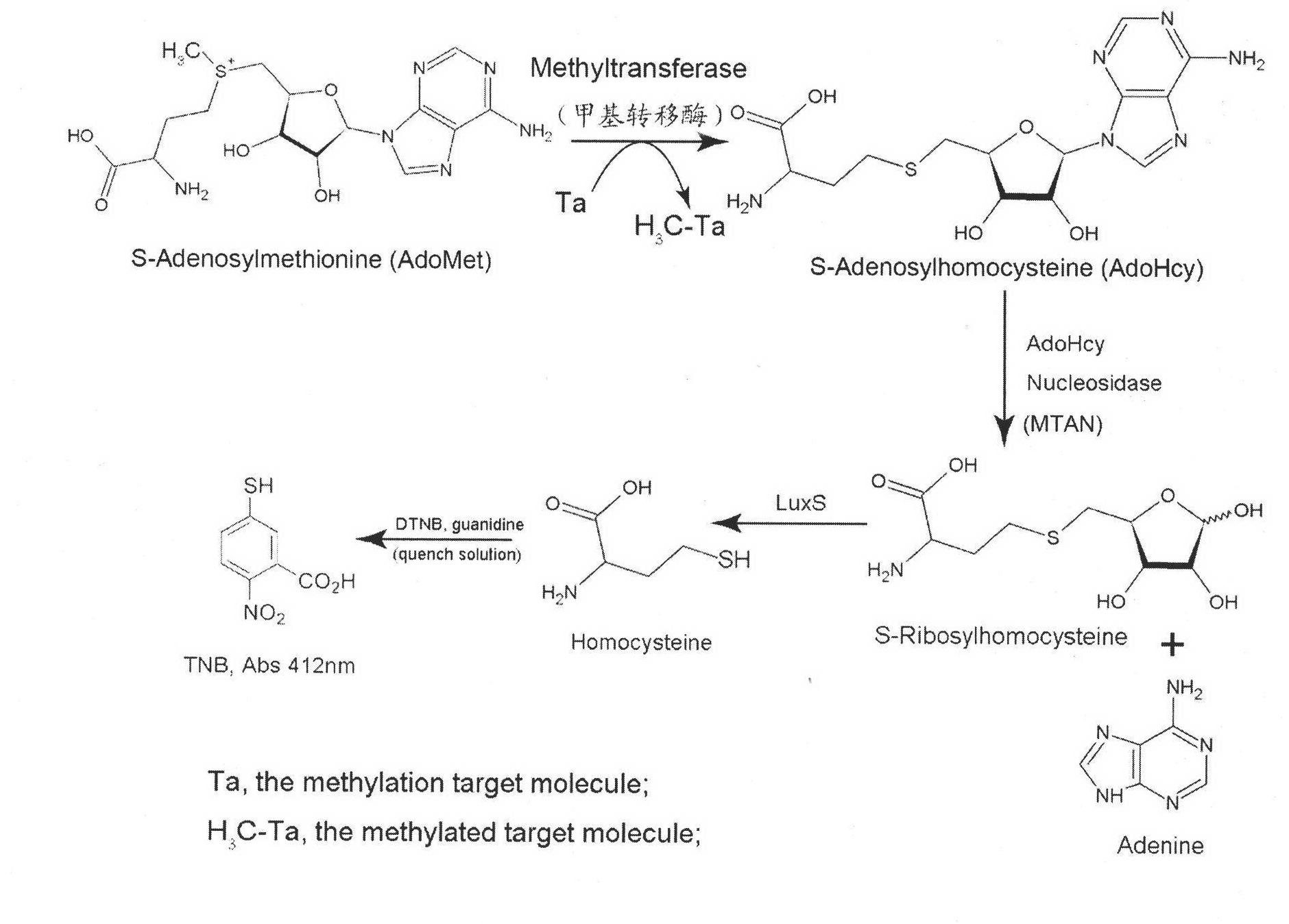
Method for detecting S-adenosylmethionine (AdoMet)-dependent methyltransferase
An adenosylmethionine, methyltransferase technology, applied in the field of biochemical analysis, can solve the problems of AdoMet's expensive price, time-consuming and laborious, affecting the accurate measurement of enzyme reaction parameters, achieve accurate and effective quantitative analysis, and eliminate feedback inhibition. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
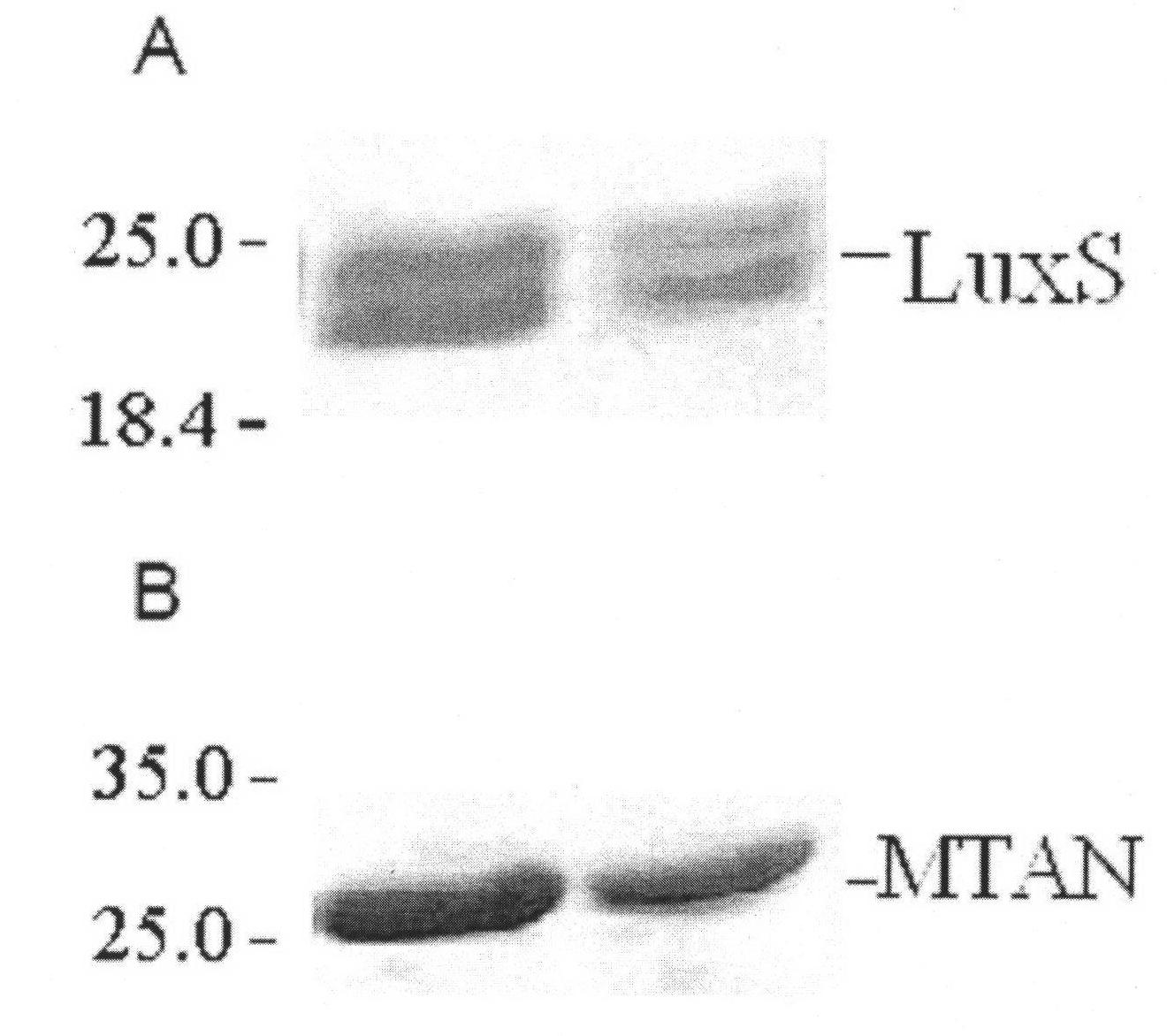
[0014] The expression and purification of recombinant S-adenosine-L-homocysteine nuclease (MTAN) includes the following steps:
[0015] i. Expression: using primers containing NdeI and BamHI endonuclease cleavage sites and using E. coli XL1-Blue chromosomal DNA as a template to amplify the 675bp polymerase chain reaction (PCR) product rMTAN- which lacks the first 8 amino acid codes 8. The PCR product was cloned into the pCRII (InVitrogen) plasmid vector, and the recombinant plasmid was transformed into competent cells of Escherichia coli TOP10F'. Alkaline lysis method prepares a small amount of recombinant plasmids from positive clones (white, ampicillin resistant), NdeI / BamHI (10 enzyme units / 37°C / 12 hours) double enzyme digestion, agarose electrophoresis, separation and purification of the digested fragments from the gel The tryptophan inducing expression vector pAL781 (InVitrogen) was cloned into the competent cell E. coli GI728, and the transformant was inoculated in Luria-...
Embodiment 2
[0018] The expression and purification of recombinant S-ribosyl homocysteinase (LuxS) includes the following steps:
[0019] i. Expression: Using Bacillus subtilis genomic DNA as a template, the LuxS gene was amplified by PCR with primers containing NdeI and NotI endonuclease cleavage sites, and the PCR product was cloned into the pET29a(+) (Novagen) expression vector to transform competent Cell E. coli BL21(DE3), recombinant plasmid pLuxSH 6 It encodes a product with 6 histidine tags at the C-terminus. The transformant was inoculated into LB broth containing 40μg / mL kanamycin and cultured with shaking at 225rpm to OD at 37°C 600 To reach 0.5-0.6, 0.4mM isopropyl-b-D-thiogalactoside (IPTG) induces the expression of S-ribosyl homocysteinase (LuxS).
[0020] ii. Purification: collect bacteria by centrifugation, wash three times with 1×PBS buffer, ultrasonic lysis centrifugation (30,000g) to remove precipitation, and load the lysate on Ni 2+ Agarose affinity chromatography column, was...
Embodiment 3
[0022] Enzyme coupling analysis based on recombinant S-adenosyl-L-homocysteine nuclease (MTAN, EC 3.2.2.9) and recombinant S-ribosyl homocysteine (LuxS, EC 3.2.1.148) The establishment of detection technology includes the following steps:
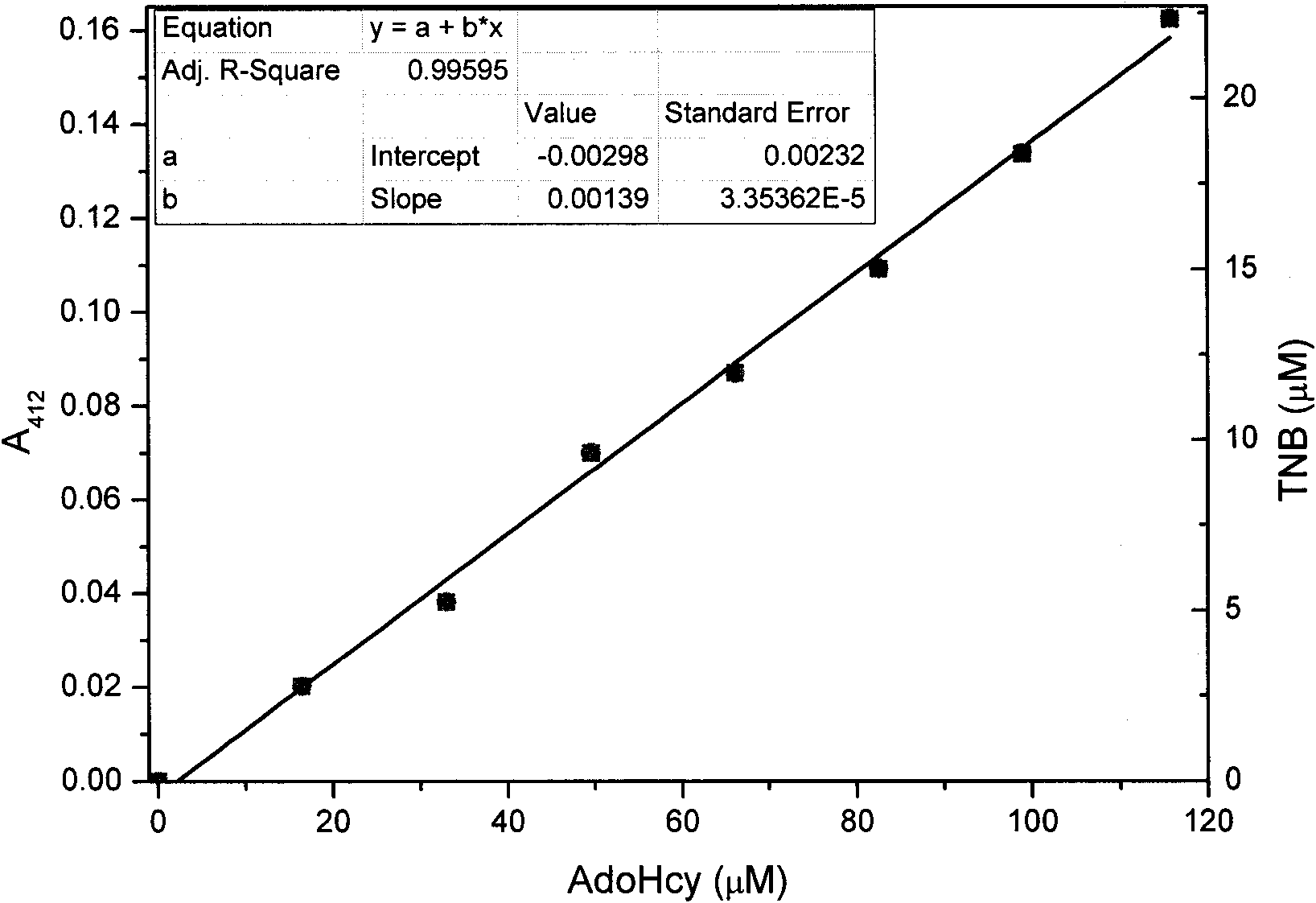
[0023] The standard curve of iS-adenosine-L-homocysteine (AdoHcy) and 5-thio-2-nitrobenzoic acid (TNB) generated by colorimetric determination: Enzyme coupling analysis detection system contains 0 , 17, 34, 51, 68, 85, 102, 119 and other different concentrations of S-adenosine-L-homocysteine (AdoHcy), and 1.5μM S-adenosine-L-homocysteine nucleoside Enzyme (MTAN), 10μM S-ribosyl homocysteinase (LuxS), 100mM Hepes buffer pH 8.0, reacted at 37°C for 30 minutes, with four times the volume of 5,5'-dithio-2,2 '-Dinitrobenzoic acid (DTNB) (133μM)-guanidine hydrochloride (8M) solution was used to terminate the reaction, and the generated 5-thio-2-nitrobenzoic acid (TNB) was measured colorimetrically at 412nm. The experimental results show ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Protein molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com