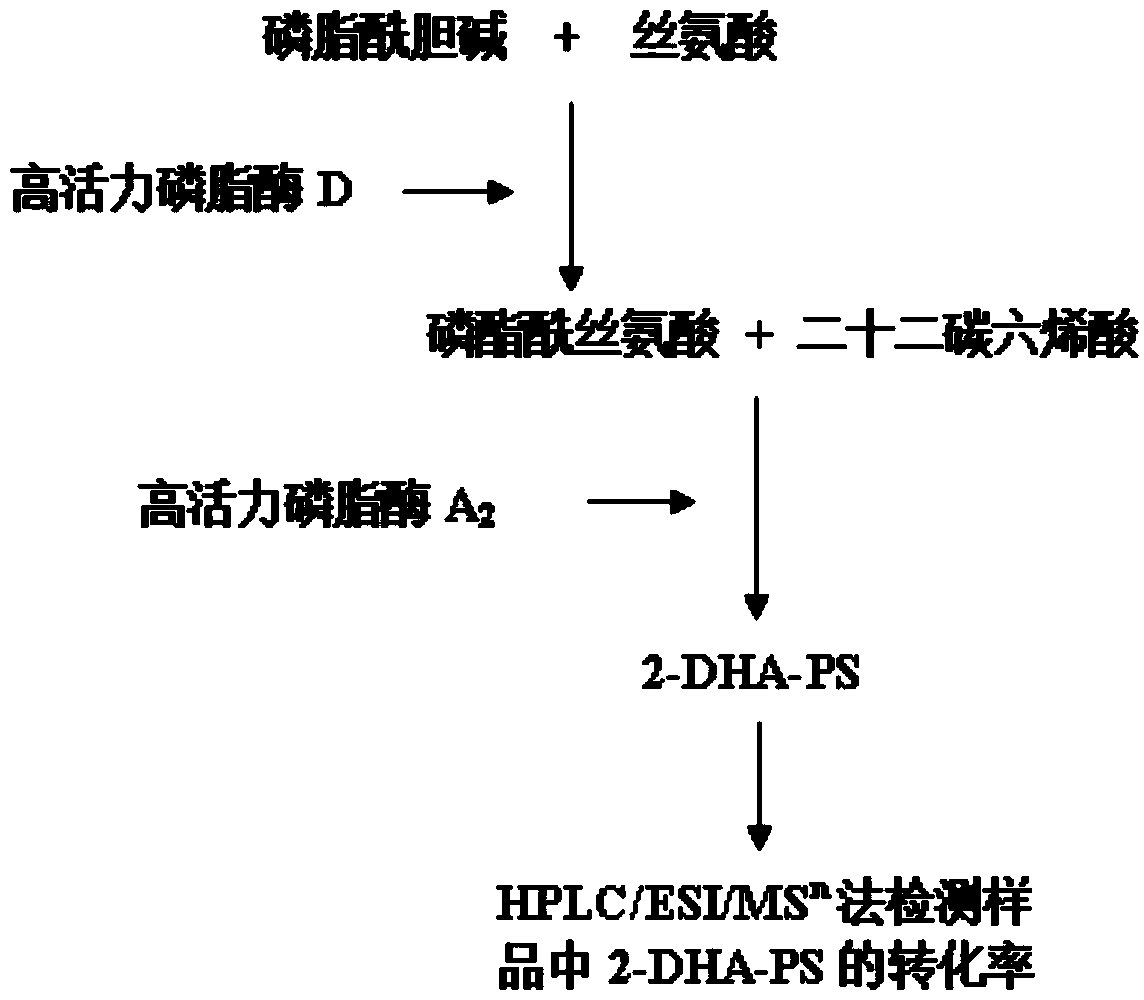
Method for preparing phosphatidylserine with docosahexaenoic acid at sn-2 bit
A phosphatidylserine and serine technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of unfavorable production and utilization of phospholipase A, narrow enzyme source, and little research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Example 1: Wild-type phospholipase A 2 Acquisition of the mature peptide gene
[0070] 1. Wild-type phospholipase A 2 The mature peptide gene comes from Streptomyces coelicolor ATCC23899, and its genomic DNA is extracted.
[0071] Wherein the extraction steps of Streptomyces coelicolor genomic DNA are as follows:
[0072] (1) Pick a ring of bacteria from the culture plate and inoculate in 40mL of appropriate medium, culture at 26°C, 150r / min for 2-3d.
[0073](2) Take 1 mL of the culture solution in a 1.5 mL EP tube, centrifuge at 12,000 r / min for 10 min, pour off the supernatant, and resuspend with 200 μL of Solution I.
[0074] (3) Add 50 μL of 50 mg / mL lysozyme and digest at 4°C for 1 hour.
[0075] (4) Add 1 / 2 volume of 2% SDS solution and react for 10 minutes until the bacterial suspension becomes viscous.
[0076] (5) Add an equal volume of saturated phenol: chloroform = 1:1, mix well, centrifuge for 10 min, transfer the supernatant to another clean EP tube, ...
Embodiment 2
[0086] Embodiment 2: high activity phospholipase A 2 Gene acquisition.
[0087] 1. Wild-type phospholipase A 2 The gene was ligated into the vector pUC-T.
[0088] Purified target gene plA 2 Ligated with vector pUC-T to form recombinant plasmid pUC-T-plA 2 , The recombinant plasmid was transformed into Escherichia coli DH5α.
[0089] 2. Site-directed mutation
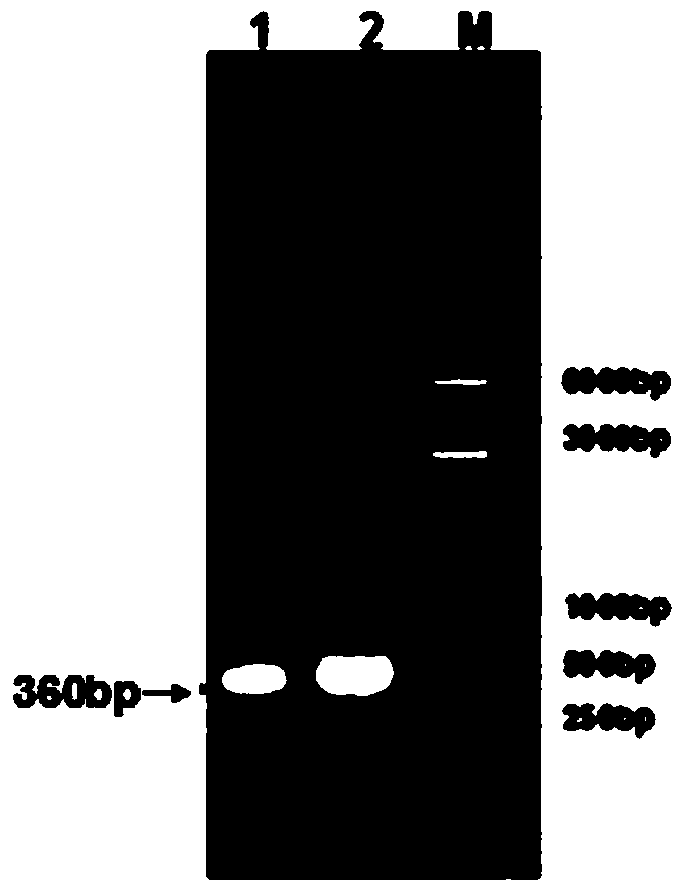
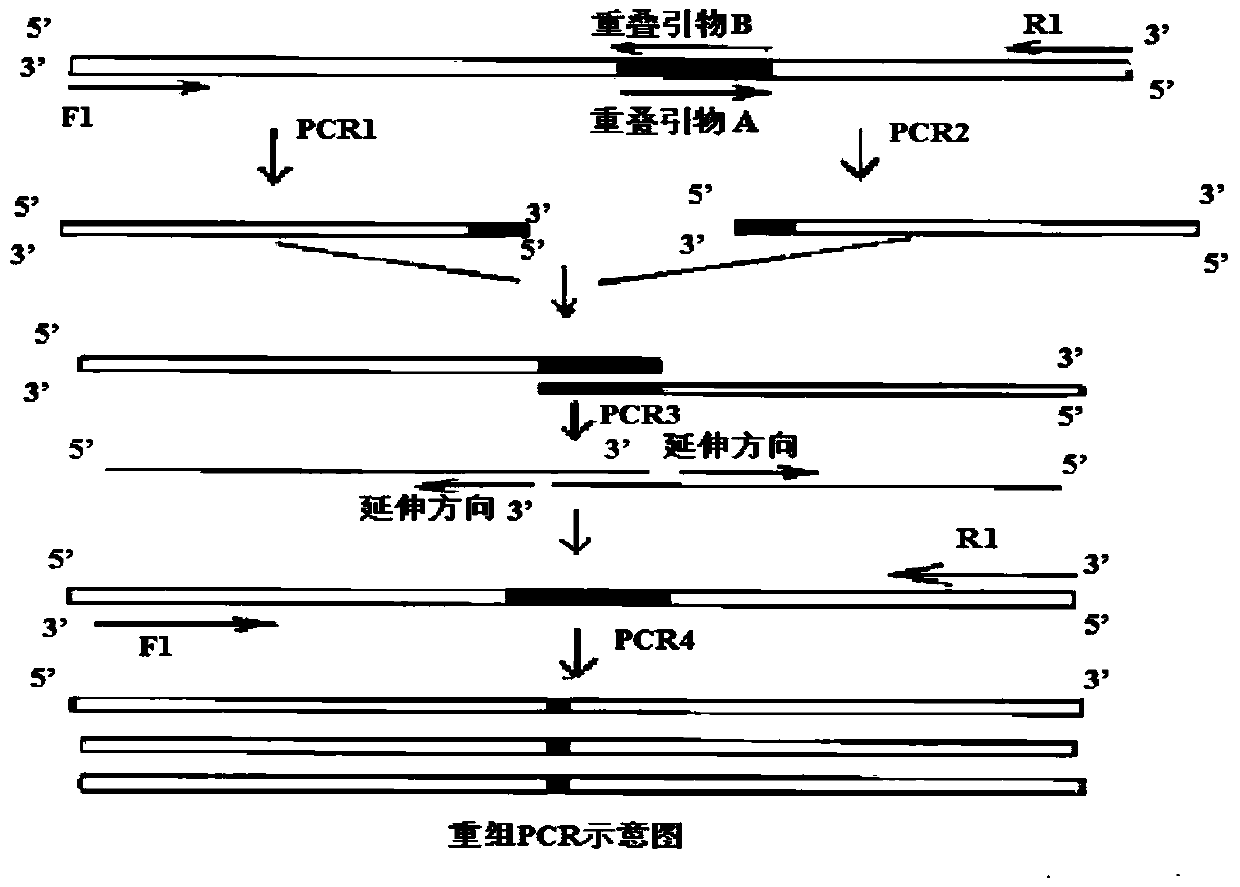
[0090] Based on overlapping PCR technology (see figure 2 ) for site-directed mutagenesis to construct a highly active phospholipase A 2 , design primers as follows:
[0091] Upstream P1 (SEQ ID NO: 1): 5'-GCCCCCGCGGACAAGCCCCAGGT-3'
[0092] Downstream P2 (SEQ ID NO: 2): 5'-TCAGCCGAAGATCTTGACGGC-3'
[0093] Overlapping primer P3 (SEQ ID NO: 3): 5'-GGCCGCCTACGCGTTCGACTGGT-3'
[0094] Overlapping primer P4 (SEQ ID NO: 4): 5'-ACCAGTCGAACGCGTAGGCGGCC-3'
[0095] Overlapping primer P5 (SEQ ID NO: 5): 5'-GGGCAGCTTCCACGCCAACAAGA-3'
[0096] Overlapping primer P6 (SEQ ID NO: 6): 5'-TCTTGTTGGCGTGGAAGCTGCCC-3'
[009...
Embodiment 3
[0125] Embodiment 3: Bacillus subtilis high activity phospholipase A 2 and the construction of highly active phospholipase D recombinant bacteria
[0126] 1. Construction of expression vector pBSA43
[0127] pBSA43 is based on the Escherichia coli-Bacillus subtilis shuttle cloning vector pBE2 as the backbone, cloned into a strong Bacillus constitutive promoter P43, and the fructan sucrase signal sequence sacB that can directly secrete the recombinant protein into the medium. get. it comes with amp r Gene that can use ampicillin resistance as a selectable marker in E. coli. At the same time with Km r , Kanamycin resistance can be used as a selection marker in Bacillus subtilis and Bacillus licheniformis.
[0128] 2. High activity phospholipase D and high activity phospholipase A 2 Expression vectors pBSA43-pldm and pBSA43-plA 2 construction of m
[0129] The high-activity phospholipase A obtained through overlapping PCR construction 2 Gene and high-activity phospholipa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com