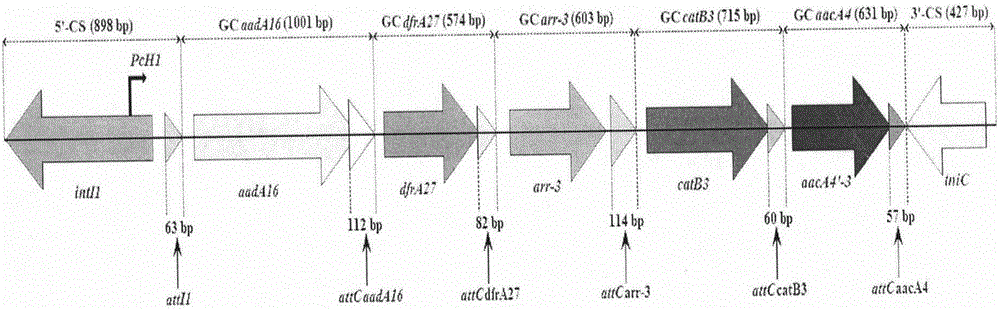
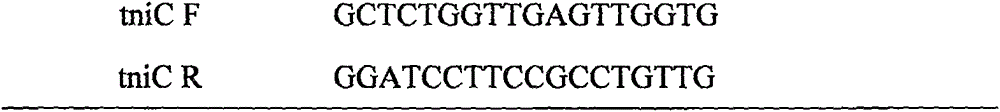Novel integron In1290
An integron, KP5325 technology, applied in the direction of biochemical equipment and methods, applications, botany equipment and methods, etc., can solve the problems of host bacteria reproduction burden and unclearness, and achieve the effect of preventing explosive epidemics and reducing selection pressure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Identification of integron In1290
[0030] 1. Isolation and identification of KP5325 strain
[0031] 1.1 Materials
[0032] Bacterial susceptibility card: AST-GN13 from bio Merieux, France. AST-GN13 drug-sensitive types include: amikacin, ampicillin, ampicillin / sulbactam, aztreonam, cefazolin, cefepime, cefotetan, ceftazidime, ceftriaxone, ciproxa Star, ertapenem, gentamicin, imipenem, levofloxacin, nitrofurantoin, piperacillin / tazobactam, tobramycin, SMZ.
[0033] Supplementary drug-sensitivity discs (drug-sensitivity plate agar diffusion test): the discs were from Oxoid, UK, and included co-trimoxazole (30 μg), chloramphenicol (30 μg) and rifampicin (5 μg).
[0034] 1.2 Method
[0035] Apparatus identification: transfer positive bacterial strains from the blood culture of patients in the Department of Infectious Diseases of Taizhou Municipal Hospital to the blood plate for isolation and culture (at 35°C with 5% CO 2 Cultivate in the incubator for 16-18h...
Embodiment 2
[0056] Example 2 Plasmid transduction experiment to study the function of integron In1290
[0057] 1. Method
[0058] (1) The donor bacterium is KP5325 strain, and the recipient bacterium is E.coli J53 Az R (resistant to sodium azide). The donor bacteria and recipient bacteria were inoculated on LB plates, respectively, and cultured overnight at 35°C. Pick a single colony and inoculate them in 4 mL of LB broth, and culture at 37°C and 220 r / min until the logarithmic growth phase. Take 0.5 mL of donor and recipient bacteria in 4 mL of LB broth, and culture overnight at 37°C. Zygotes were screened on trypan soy agar (TSA) plates containing sodium azide (300 mg / L) and amifloxacin (0.06 mg / L). Incubate at 35°C for 18-24h. The plasmid of the drug-resistant strain was extracted (the steps are the same as in Example 1), and the In1290 sequence of the integron was detected by the PCR method.
[0059] The primers for amplifying the integron In1290 sequence are shown in Table 1: ...
Embodiment 3
[0070] Example 3 Gene recombination experiments to study the function of integron In1290
[0071] 1. Method
[0072] (1) Ligate the PCR product in Example 2 (the nucleotide sequence is SEQ ID NO.1) to the pMD19-T vector, add 100 μL of E.coli JM109 to the competent state of the ligation product, and transform it in a culture medium containing IPTG, x- The gal and Amp plates were cultured, the IPTG, x-gal, and Amp resistant strains were extracted and the plasmids were sequenced (the steps were the same as in Example 1), and it was detected whether the integron In1290 was inserted into the genome. Then the integron In1290 sequence was cloned into the pET32a vector, the insertion site of the integron was at the BamHI restriction site on the pET32a vector, prepared according to conventional methods, and the pET32a recombinant plasmid containing the integron In1290 sequence was obtained. Transform the recombinant plasmid into competent E.coli JM109 cells.
[0073] (2) Detection of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com