Polymerase chain reaction primers for quantitatively detecting Ochrobactrum anthropi, kit and method for quantitatively detecting Ochrobactrum anthropi
A paleobacterial, quantitative detection technology, applied in microorganism-based methods, biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of errors, high cost, slow speed, etc. Detecting fast effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1: the verification of paleobacterium microorganism-specific primer pair
[0045] The designed primer pair OA3F-OA3R was used for PCR amplification experiment. The PCR reaction system (12 μL) was: NPK02buffer (2×) (Weihai Xiaodong Biology) 6 μL, species-specific primers 1.5 μL (including both upstream and downstream primers, concentration 2 μM), template 1 μL (metagenomic DNA extracted from pit mud soil samples and marine biofilm samples as template DNA), Taq enzyme 0.25 μL, dd H 2 O 3.25 μL.
[0046] PCR reaction cycle parameters: 94°C for 3min; 94°C for 30s, 60°C for 30s, 72°C for 1min, 37 cycles; 72°C for 2min.
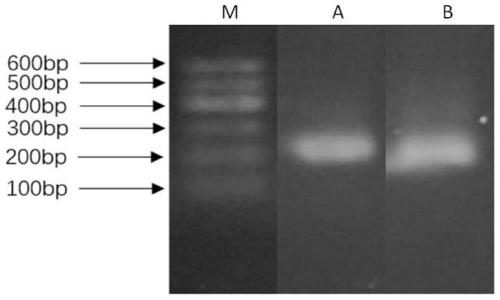
[0047] Agarose gel electrophoresis: 1% agarose gel, 4 μL of MarkerB sample, 12 μL of amplification product + 3 μL of loading buffer, and an appropriate amount of ethidium bromide (10% EB) was added to the buffer. The voltage is 110V, the current is above 40mA, observe once every 20min, and stop electrophoresis for about 40min.
[0048] The spec...
Embodiment 2
[0063] The specificity of the DNA fragment detected in embodiment 2 in other pale bacilli
[0064] The OA3F-OA3R primers were used to amplify the sequence of Paleobacterium pallidum in the rice field sludge metagenome, and the PCR product was sequenced using OA3R unidirectionally, and the sequencing results were converted into reverse complementary sequences (forward sequence), the results are as follows:
[0065] >1
[0066] AACGGGAACCCGTCGAAGAACGAGCCGCCGGCGAACAGCACGATCAGCAGGATGGCGAACAGGAAGCTCGGAATGGCGTAGCCGACGATGATGACGGCGCTGGTCCAGGTGTCGAAGCGCGAGCCGTCCTTGATCGCCTTGCGGATGCCGAGCGGGATCGAGATCAGGTAAGGGACAGCAAGGTCGAACCAAGGCCCGAAC 201 bases
[0067] Expected sequence:
[0068] >2
[0069] CAACGGGAACCAGTCGAAGAACGAGCCGCCCGCAAACAGTACGATCAGCAGGATTGCAAACAGGAAGCTTGGAATGGCGTAGCCGACGATGATGACGGCGCTGGTCCAGGTGTCGAAGCGCGAACCGTCCTTGATTGCCTTGCGAATGCCGAGTGGAATCGAGATCATGTAGGACAGAAAAGTCAGCCACAGGCCGAGCGAGATTGAAACCGGCATC 217碱基(Ochrobactrum anthropi strain OAB chromosome 1,Range:263890to 264106)(Ochro...
Embodiment 3
[0073] Example 3 Using OA3F-OA3R Primers to Quantitatively Determinate the Content of Paddy Paddy Soil Samples
[0074] In this embodiment, the compound bacteria preparation contains Pediococcus acidilactici, Lactobacillus plantarum (Lactobacillus plantarum) Lactobacillus plantarum, Lactobacillus pentosus lactobacillus pentosus, Lactobacillus paracasei Lactobacillus paracasei, Agrobacterium tumefaciens, Serrat Serratia sp.JL, Lactobacillus fermentum, Agrobacterium Rhizobium larrymoorei, iron-based autotrophic nitrifying bacteria Microbacterium testaceum, Luteimonas cucumeris, Ochrobactrum sp.JL, Lactobacillus casei, nasturtium Fructobacillus tropaeoli, Acetobacter setunensis.
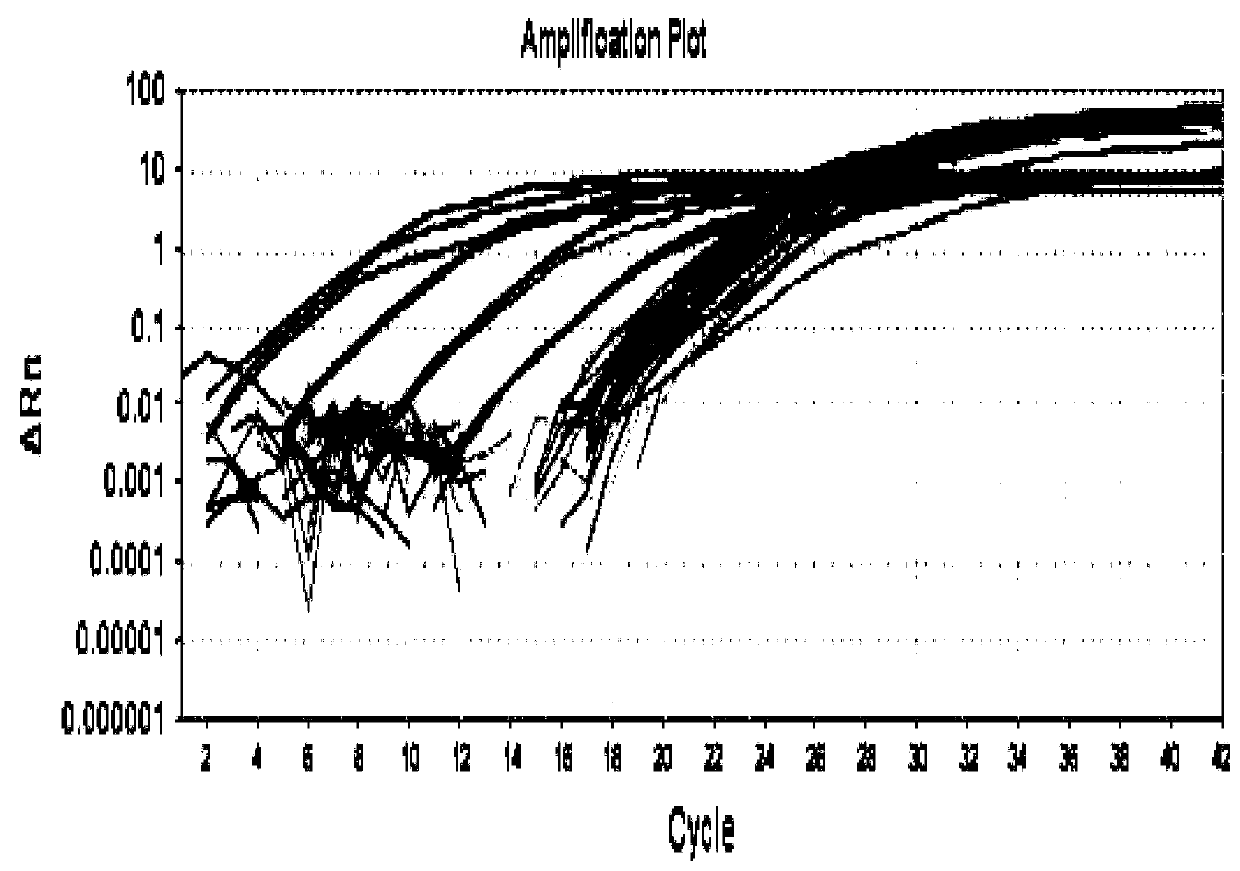
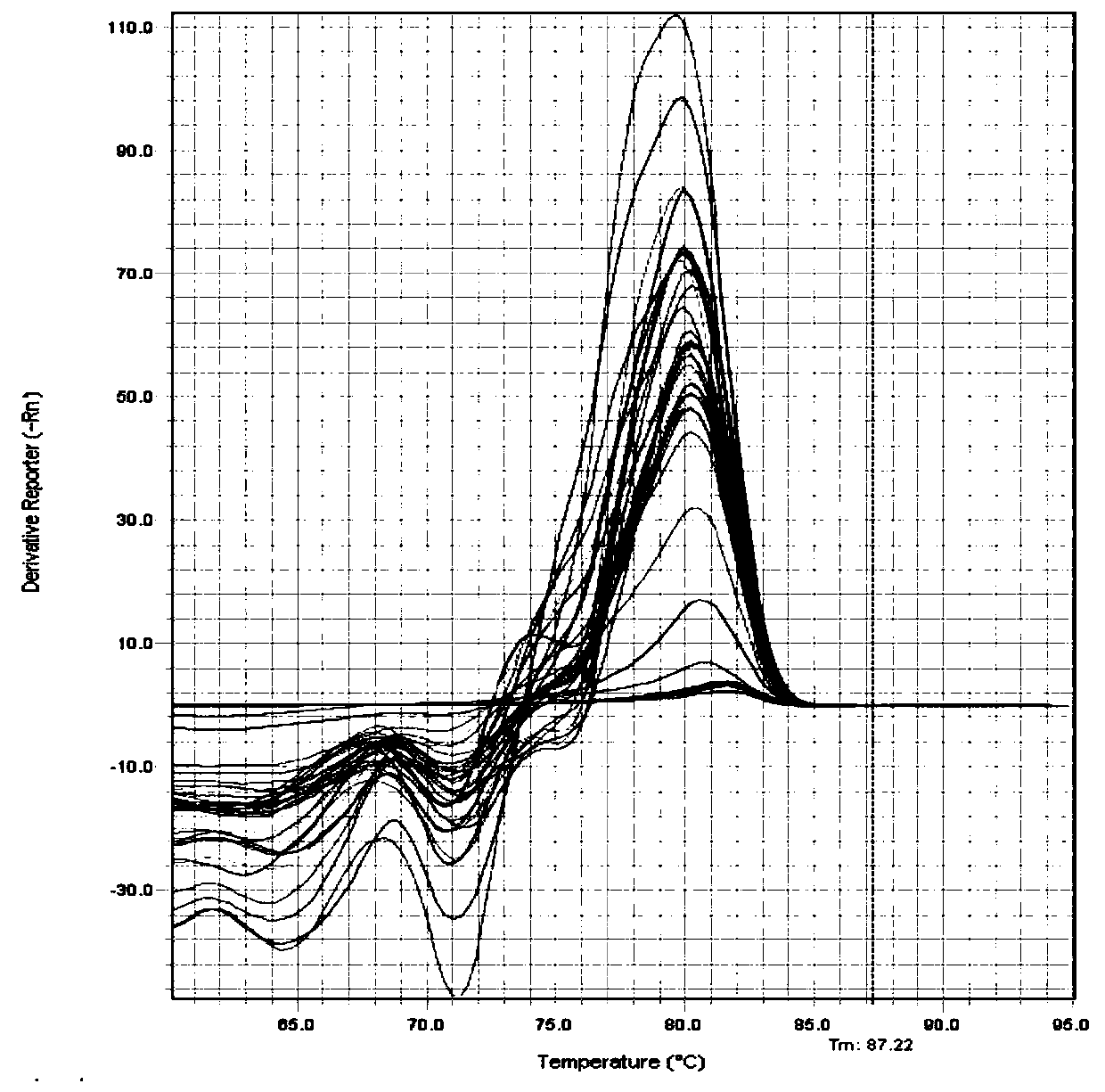
[0075]QPCR determination of Pallidum pallidum in a series of paddy field soil samples: Two soil samples with and without complex bacterial preparations were extracted by conventional methods. Use the Ezup column genome extraction kit (B518251, Shanghai Sangong) to extract 32 samples (including 16 gener...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com