Novel method for integrating genes at specific sites in mammalian cells via homologous recombination and vectors for accomplishing the same
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Design and Preparation of Marker and Targeting Plasmid DNA Vectors
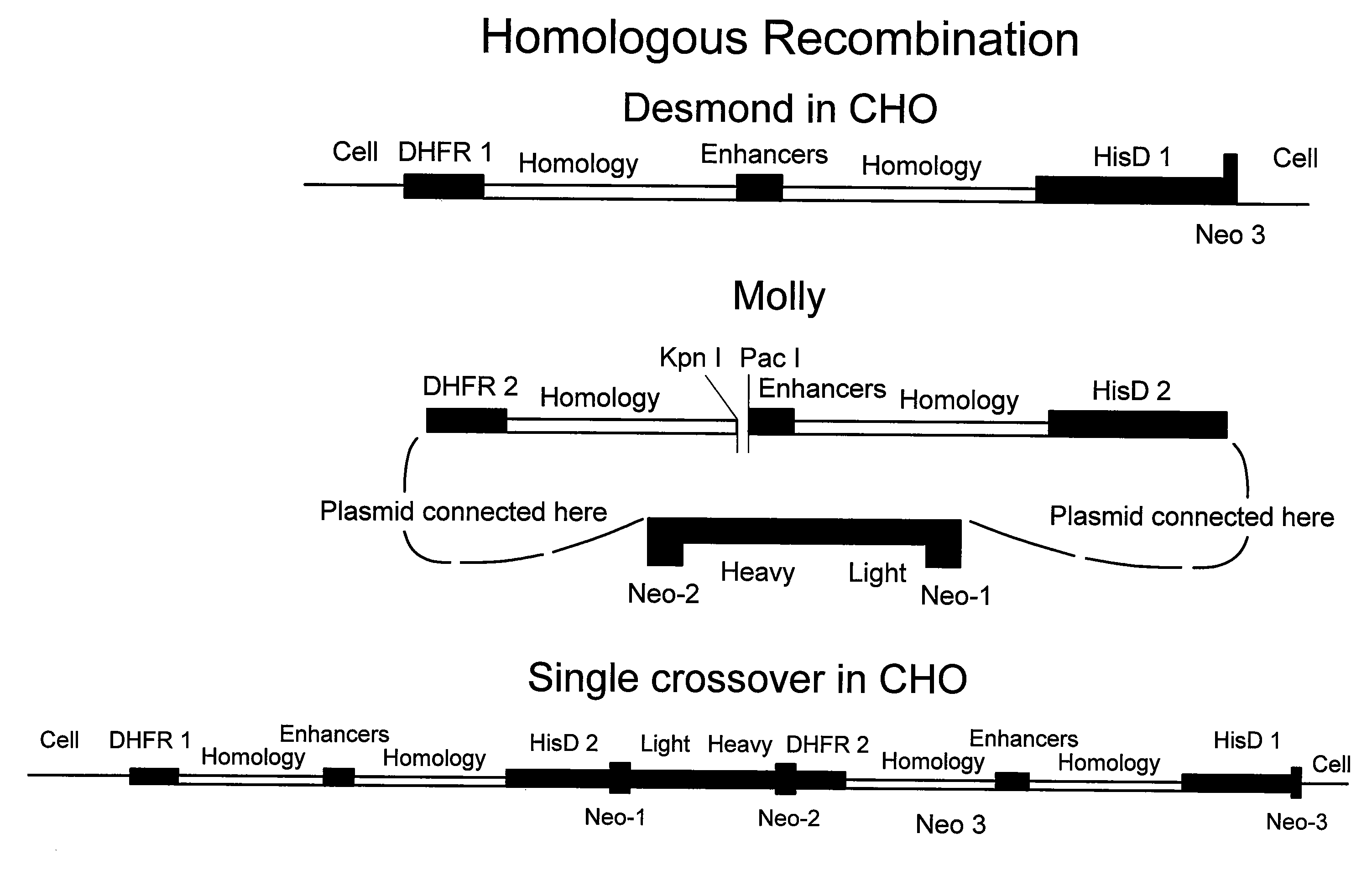
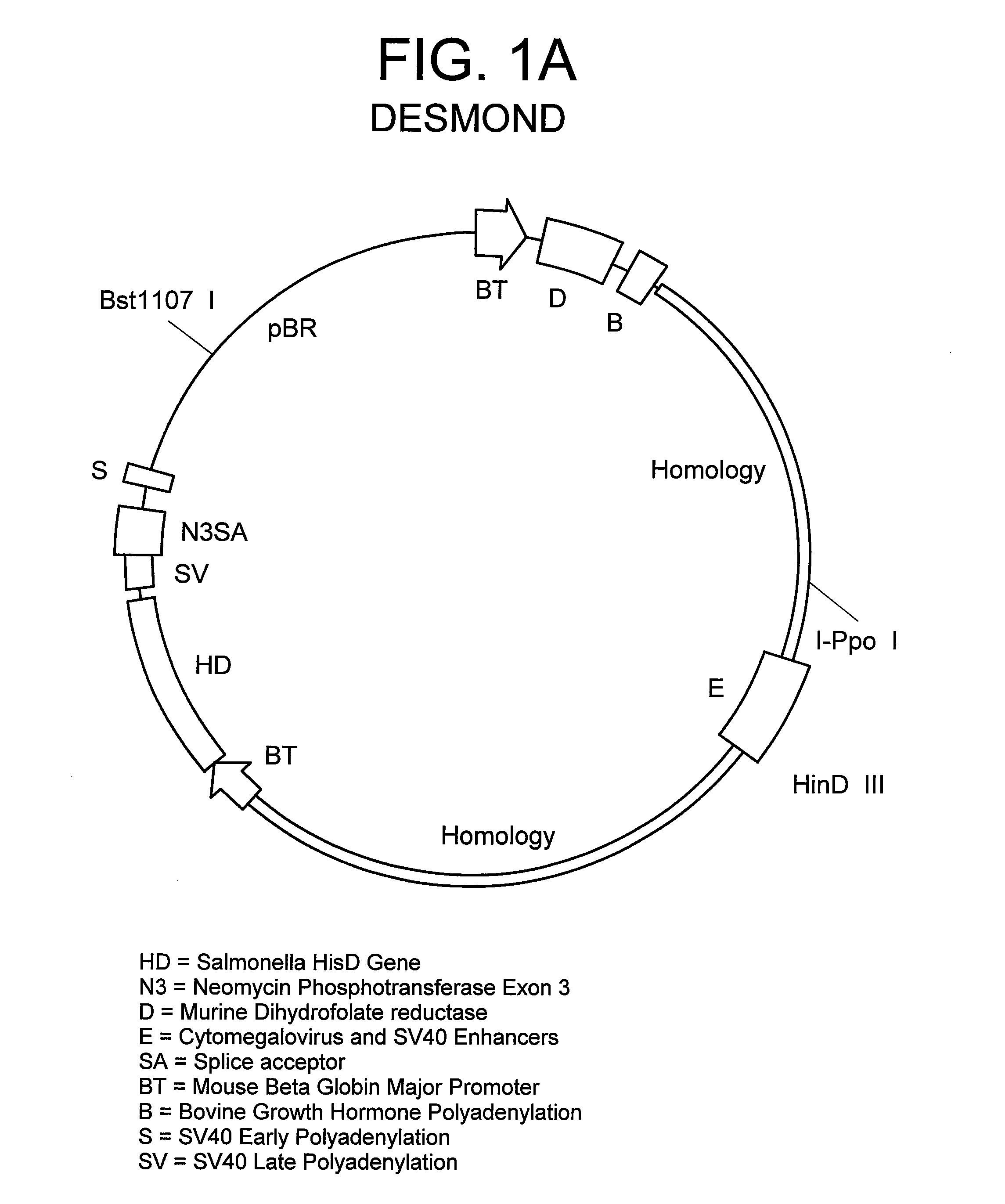
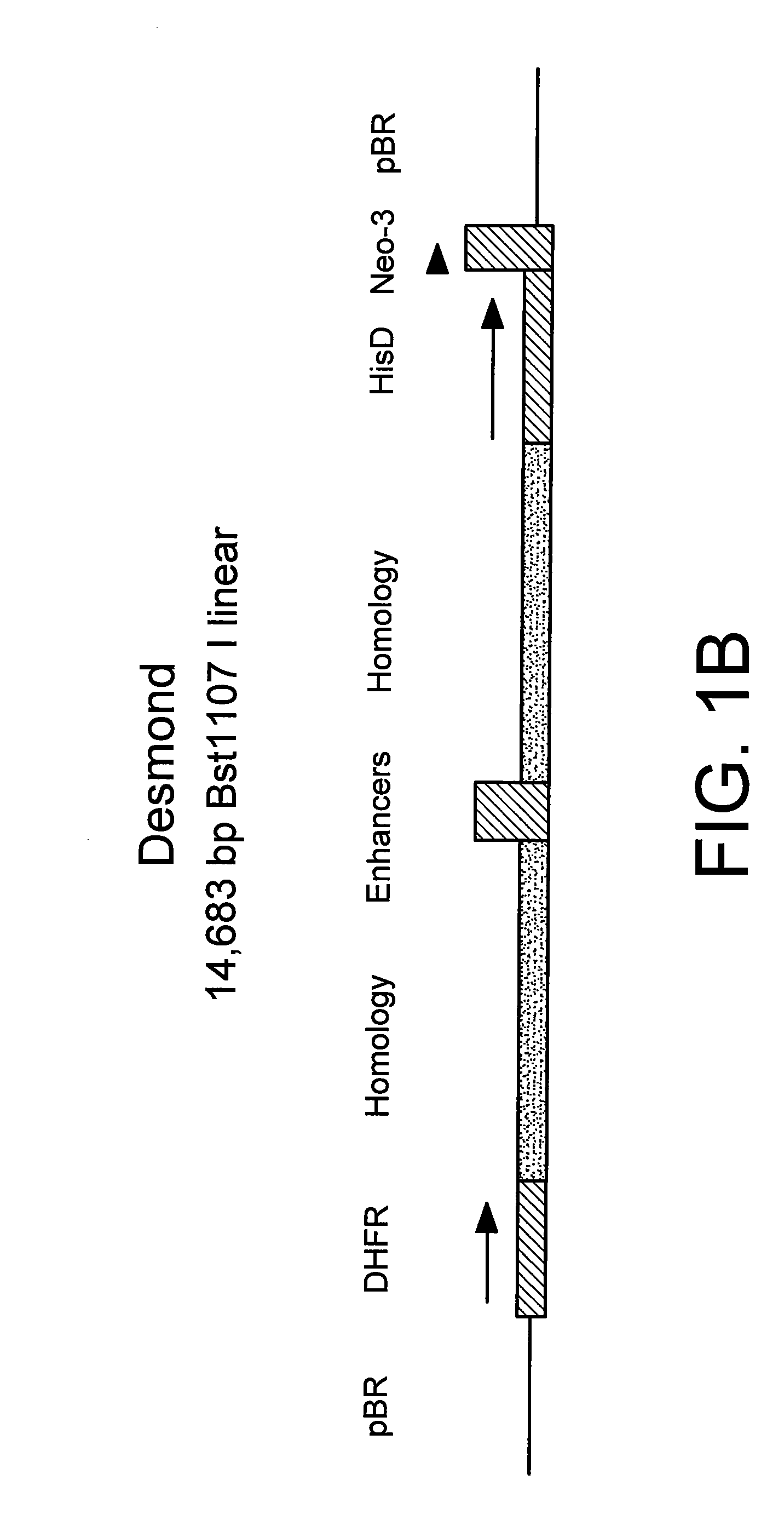
[0069]The marker plasmid herein referred to as “Desmond” was assembled from the following DNA elements:
[0070](a) Murine dihydrofolate reductase gene (DHFR), incorporated into a transcription cassette, comprising the mouse beta globin promoter 5″ to the DHFR start site, and bovine growth hormone poly adenylation signal 3″ to the stop codon. The DHFR transcriptional cassette was isolated from TCAE6, an expression vector created previously in this laboratory (Newman et al, 1992, Biotechnology, 10:1455-1460).
[0071](b) E. coli β-galactosidase gene—commercially available, obtained from Promega as pSV-b-galactosidase control vector, catalog # E1081.
[0072](c) Baculovirus DNA, commercially available, purchased from Clontech as pBAKPAK8, cat # 6145-1.
[0073](d) Cassette comprising promoter and enhancer elements from Cytomegalovirus and SV40 virus. The cassette was generated by PCR using a derivative of expression vector TCAE8 (R...
example 2
Construction of a Marked CHO Cell Line
1. Cell Culture and Transfection Procedures to Produced Marked CHO Cell Line
[0086]Marker plasmid DNA was linearized by digestion overnight at 37° C. with Bst1107I. Linearized vector was ethanol precipitated and resuspended in sterile TE to a concentration of 1 mg / ml. Linearized vector was introduced into DHFR-Chinese hamster ovary cells (CHO cells) DG44 cells (Urlaub et al, Som. Cell and Mol. Gen., 12:555-566 (1986)) by electroporation as follows.
[0087]Exponentially growing cells were harvested by centrifugation, washed once in ice cold SBS (sucrose buffered solution, 272 mM sucrose, 7 mM sodium phosphate, pH 7.4, 1 mM magnesium chloride) then resuspended in SBS to a concentration of 107 cells / ml. After a 15 minute incubation on ice, 0.4 ml of the cell suspension was mixed with 40 μg linearized DNA in a disposable electroporation cuvette. Cells were shocked using a BTX electrocell manipulator (San Diego, Calif.) set at 230 volts, 400 microfarada...
example 3
Characterization of Marked CHO Cell Lines
(a) Southern Analysis
[0088]Genomic DNA was isolated from all stably growing Desmond marked CHO cells. DNA was isolated using the Invitrogen Easy® DNA kit, according to the manufacturer's directions. Genomic DNA was then digested with HindIII overnight at 37° C., and subjected to Southern analysis using a PCR generated digoxygenin labelled probe specific to the DHFR gene. Hybridizations and washes were carried out using Boehringer Mannheim's DIG easy hyb (catalog # 1603 558) and DIG Wash and Block Buffer Set (catalog # 1585 762) according to the manufacturer's directions. DNA samples containing a single band hybridizing to the DHFR probe were assumed to be Desmond clones arising from a single cell which had integrated a single copy of the plasmid. These clones were retained for further analysis. Out of a total of 45 HisD resistant cell lines isolated, only 5 were single copy integrants. FIG. 4 shows a Southern blot containing all 5 of these si...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com