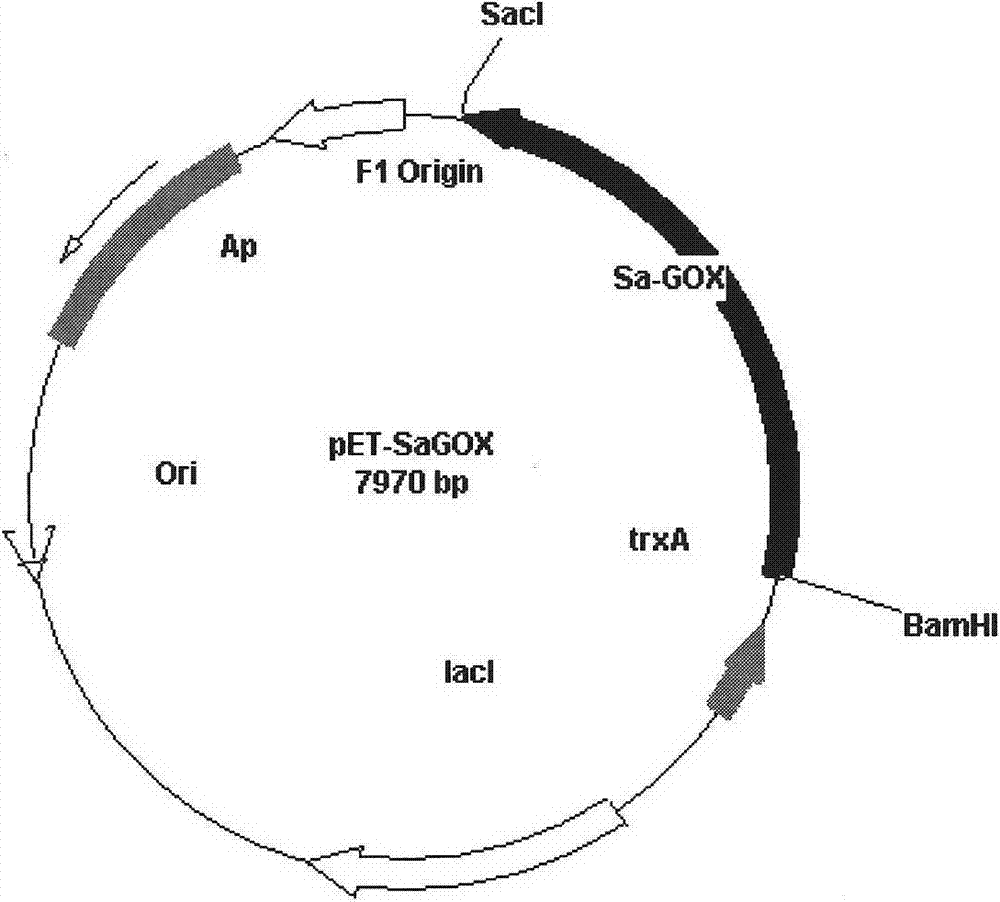
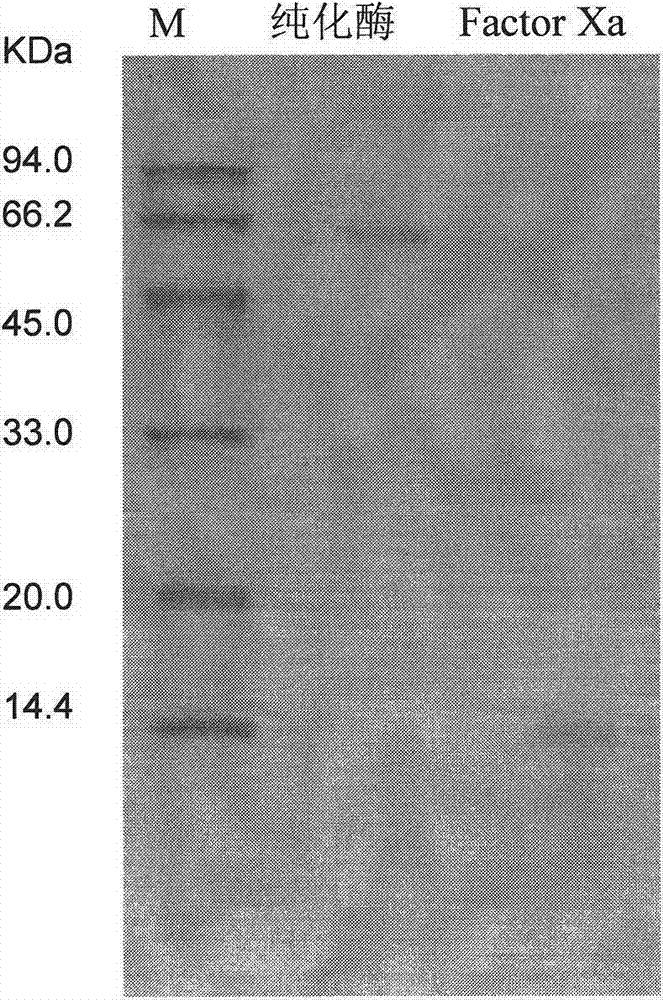
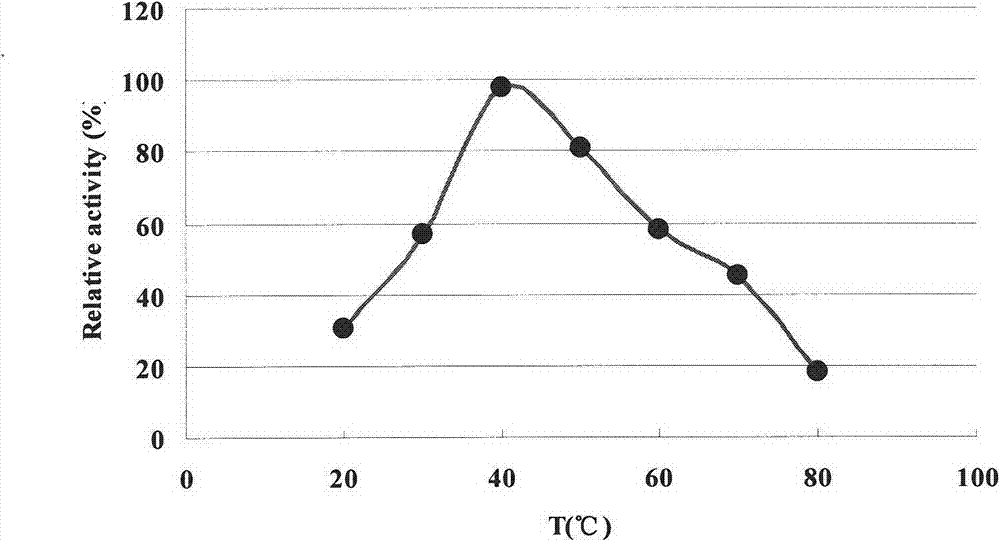
L-glutamate oxidase gene from Afghanistan streptomyces and application thereof
A technology of glutamate oxidase and Streptomyces, applied in application, genetic engineering, plant genetic improvement and other directions, can solve the problems of low enzyme production capacity and low enzyme production capacity, and achieve high specificity and reduce extraction costs. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1, strain separation
[0036] Weigh 1 g of soil sample, add 1 ml of 0.9% (w / v) sodium chloride solution, shake and mix at 5000 rpm, centrifuge gently at 3000 rpm, pour off the supernatant, and then add 0.9% (w / v) chlorine Sodium chloride solution 1ml, shake and mix at 5000 rpm, let it stand on ice for 10min, draw 100μL solution and spread it on Gaoshi No. 1 medium containing 50ppm potassium dichromate, 30 ℃ cultured for 4 days. According to the shape and size of the colonies, the colonies were picked and respectively inserted into the control and color development plates (both in the plates, Gaoshi No. 1 medium), 30 ℃ Cultivate for 3 days under low temperature, take the color developing plate, put the filter paper soaked in the color developing solution on the colony, mark the colony with fast color development and deep red color on the control plate corresponding to the colony, pick the colony and cultivate it in Gaoshi No. 1 Separation on dashed base, 30 ...
Embodiment 2
[0037] Embodiment 2, total DNA extraction and strain identification
[0038] The isolated bacterial strain was inoculated in 20mL liquid medium (yeast powder 3g / L, peptone 20g / L, glucose 10g / L, soluble starch 5g / L, soybean peptone 5g / L, MgSO 4 ·7H 2 (00.2g / L, pH=7.0), 30 ℃ After 3 days of cultivation, the culture solution was centrifuged at a speed of 8000r / min for 5 minutes to obtain bacterial pellets.
[0039] Take 0.15g of fresh bacteria in a mortar (-20 ℃ pre-cooled), ground repeatedly with liquid nitrogen until fine powder, quickly put into a 1.5mL centrifuge tube, added 550μL of TE buffer, 65 ℃30 μL of preheated 20% (w / v) SDS solution, mediate and shake for 5 minutes, then add 20 μL of proteinase K with a concentration of 20 mg / mL, mix well, bathe in water at 37°C for 1 hour, add an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1, v / v) extraction, gently inverted and mixed, centrifuged at 10000r / min for 10 minutes; pipette the supernatant into a new cent...
Embodiment 3
[0043] Embodiment 3, acquisition of the L-glutamate oxidase gene of Streptomyces afghanistan
[0044] Primers were designed according to the conserved box sequence of L-glutamic acid oxidase: Primer A: 5'-AGTACGCGGAGGCCGGCGCCAT-3' and Primer B: 5'-GCTGAACTCCAGCAGCACCTTGGT-3' as amplification primers, using the total DNA as a template to obtain a 984bp PCR fragments. The PCR reaction conditions were: 94°C for 30s, 55°C for 30s, 72°C for 90s, and 30 cycles of amplification. The fragment was recovered with 1% agarose gel, and 10 μl was directly connected to the T / A cloning vector (Dalian Bao Biological Company). 4 ℃ After ligation overnight, high-efficiency transformation in DH5α competent cells, positive clones were obtained and sequenced. Based on this sequence, the Genome Walking method was used to obtain the unknown sequences on both sides, and the complete open reading frame was searched, so as to obtain the complete sequence of the L-glutamic acid oxidase gene.
[0045]...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Final concentration | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com