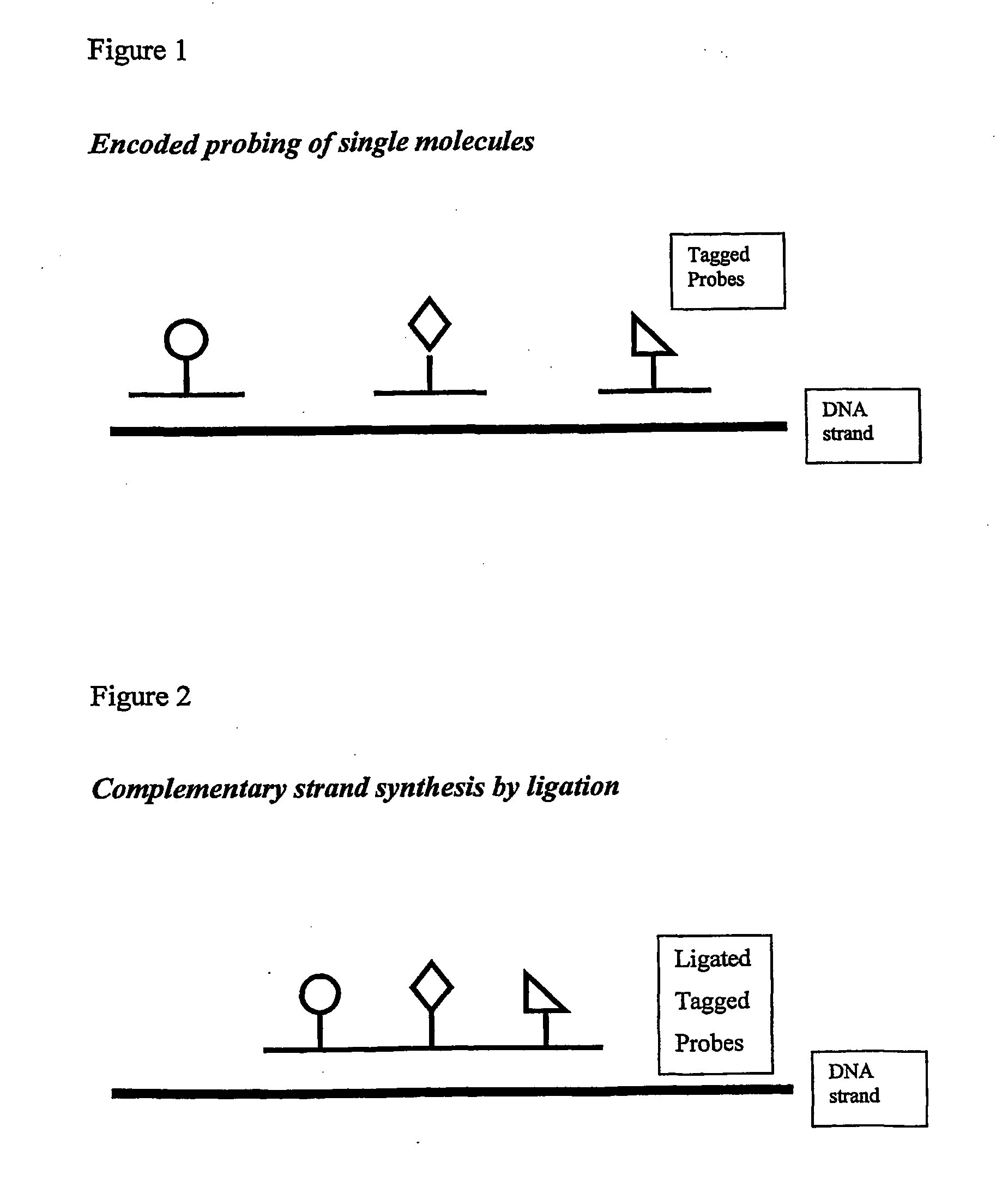
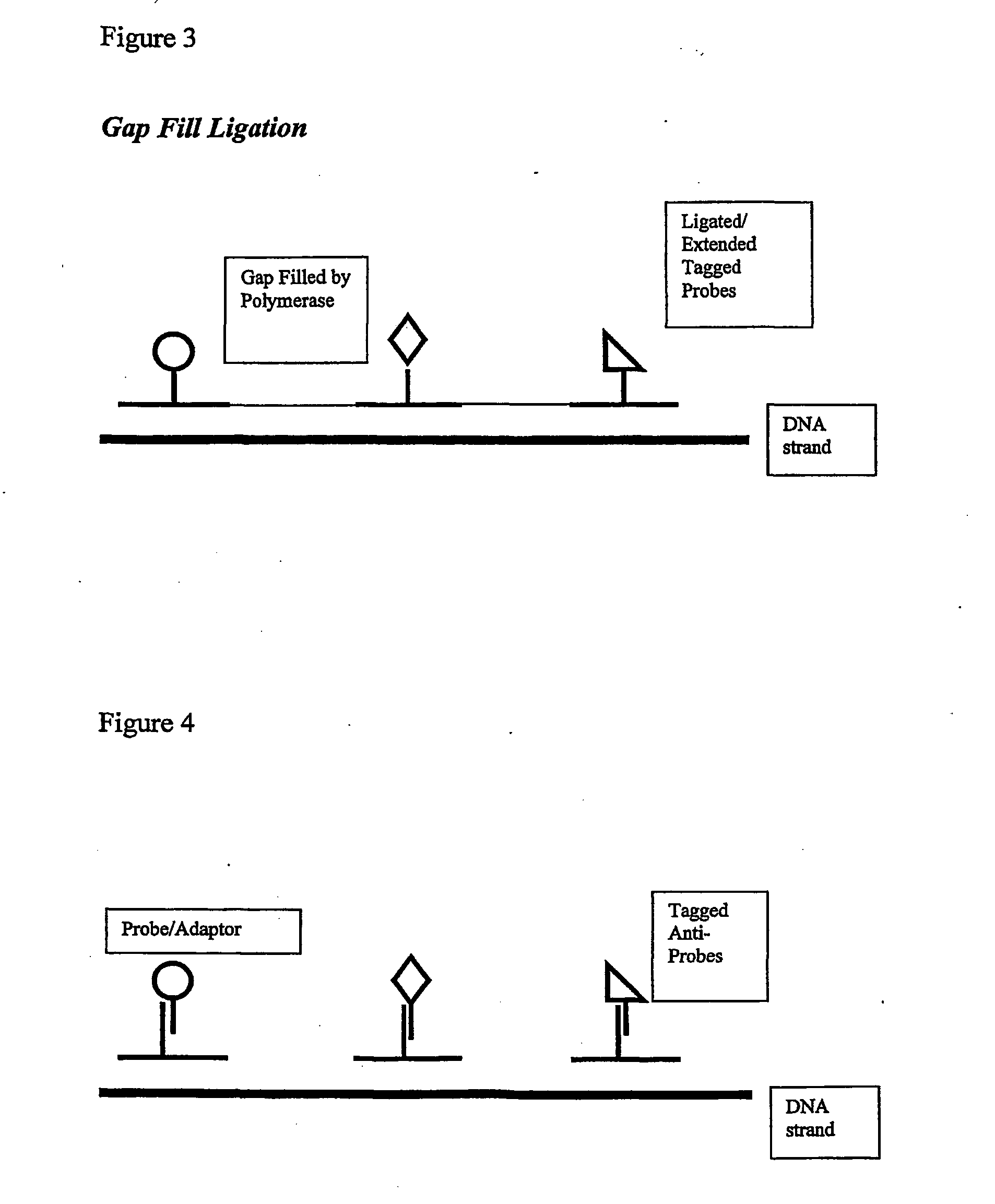
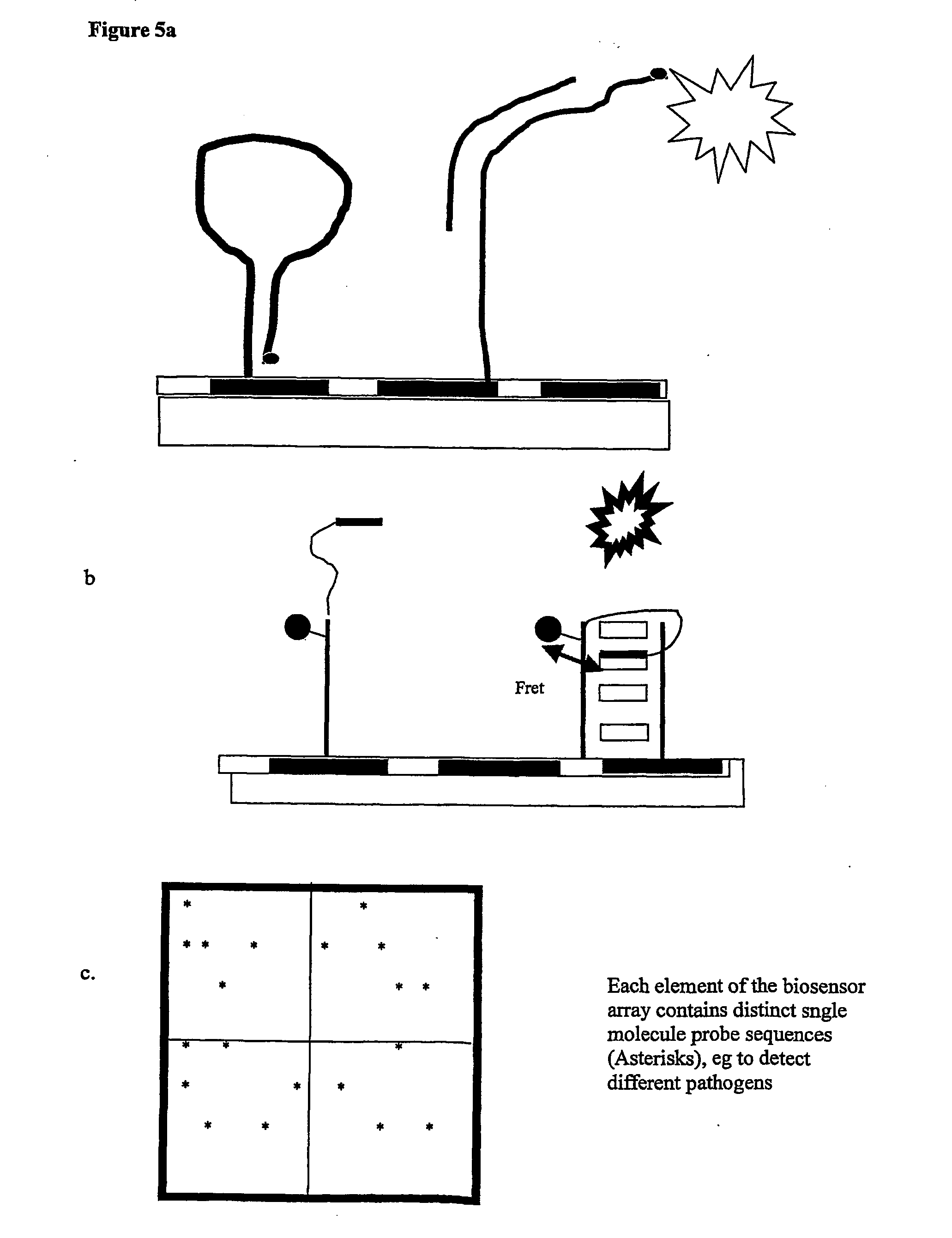
Although bulk or ensemble approaches have in the past proved useful, there are barriers to progress in a number of directions.
The practical limitations associated with bulk analysis include the following:
1. The techniques used for the detection of events in bulk
phase analysis are not sensitive enough to detect rare events which may be due to low sample amount or
weak interaction with probes.
This problem is related to the limited
dynamic range of bulk analysis which is in the order of 10.sup.4 whereas the different abundance levels of mRNAs in a
cell are in the 10.sup.5 range.
Hence to cater for the more common events, detection methods are not sensitive enough to detect rare events.
b. In the amounts of samples that are usually available to perform
genetic analysis there are not enough copies of each sequence in
genomic DNA to be detected.
These events may be too few to be detected by conventional bulk measurements.
In analysis of ancient
DNA the amount of sample material available is oftenalso very small.
There are a number of instances where this is important:
a. Detecting loss of heterozygosity (LOH) in tumours comprising
mixed cell populations and early events in tumourigenesis.
c.
Prenatal diagnosis of genetic disorders directly from the small number of foetal cells in the maternal circulation (hence detection from mother's blood rather than from amniocentesis).
d. Detection of specific alleles in pooled
population samples.
3. It is difficult to resolve heterogeneous events. For example it is difficult to separate out the contribution (or the lack of) to
signal from errors such as foldback, mis-priming or self-priming from genuine signals based on the interactions being measured.
4. Complex samples such as
genomic DNA and mRNA populations
pose difficulties.
b. On arrays, Another is the high degree of erroroneous interactions which in many cases are likely to be due to mismatch interactions driven by high effective concentrations of certain species. This is one reason for low
signal to
noise. A ratio as low as 1:1.2 has been used in published array studies for
base calling (Cronin et al, Human
Mutation 7:244-55, 1996).
c. In some cases erroneous interactions can even be responsible for the majority of
signal (Mir, K; D. Phil thesis, Oxford University, 1995).
d. Detecting a true representative signal of a rare mRNA transcript within a mRNA
population is difficult.
5. The bulk nature of conventional methods does not allow access to specific characteristics (particularly, more than one feature) of individual molecules. One example in
genetic analysis is the need to obtain genetic phase or
haplotype information--the specific alleles associated with each
chromosome. Bulk analysis cannot resolve
haplotype from a heterozygotic sample. Current
molecular biology techniques that are available, such as
allele-specific or single molecule PCR are difficult to optimise and apply on a large scale.
6. Transient processes are difficult to resolve. This is needed when deciphering the molecular mechanisms of processes. Also transient
molecular binding events (such as
nucleation of a hybridisation event which is blocked from propagation due to secondary structure in the target) have fractional occupancy times which cannot be detected by conventional
solid-phase binding assays.
When two samples are compared, small differences in concentration (less than twofold difference) are difficult to unequivocally discern.
The need to design primers and perform PCR on a large number of SNP sites presents a major drawback.
The largest scales of analysis that are currently being implemented (e.g. using Orchid Bioscience and Sequenom systems) remain too expensive to allow meaningful association studies to be performed by all but a few large organizations such as the Pharmaceutical companies.
Even so, if each site had to be amplified individually the task would be enormous.
However, the extent to which this can be done is limited and increased errors, such as primer-
dimer formation and mismatches as well as the increased
viscosity of reaction, present barriers to success and limits
multiplexing to around ten sites in most laboratories.
It is clear that the cost of performing SNP detection reactions on the scale required for high-
throughput analysis of polymorphisms in a
population is prohibitive if each reaction needs to be conducted separately, or if only a limited
multiplexing possibility exists.
DNA pooling is a solution for some aspects of genetic analysis but accurate
allele frequencies must be obtained which is difficult especially for rare alleles.
However practical use of this set is confounded by the fact that different SNPs may be common in different ethnic populations and many of the putative SNPs may not be truly polymorphic.
Furthermore, the CD / CV
hypothesis has recently come under challenge from assertions that rare alleles may contribute to the common diseases (Weiss K M, Clark A G, Trends Genet 2002 January;18(1):19-24).
This cost and timescale is prohibitive as an alternative to SNP analysis for finding associations between DNA sequence and
disease.
However, the cost of large scale re-sequencing by this method is still high and only 65% of the bases that were probed gave results of enough confidence for the base to be called.
To date single molecule analysis has only been conducted in simple examples but as mentioned above the challenge of modern
genetics and other areas is to apply tests on a large scale.
The
low density signals from these arrays may not be sufficiently readable by
instrumentation typically used for analysing the results of bulk arrays particularly due to high background.
Thus, there is no requirement to amplify target nucleic acids, which is a very cumbersome task when analysis is large scale or requires rapid turnaround and which can introduce errors due to non-
linear amplification of target strands and the under-representation of rare molecular species often encountered with PCR.
Low signal intensities reduce the accuracy with which the spatial position of a single molecule can be determined.
The use of dye molecules encounters the problems of
photobleaching and blinking.
Microscopy and array scanning are not typically configured for single molecule detection.
However, the problem is not so much the detection of
fluorescence from the desired single molecule (single fluorophores can emit .about.10.sup.8 photons / sec) but the rejection of
background fluorescence.
It can be difficult to differentiate between correct incorporation and mis-incorporation in the mini-sequencing (multi-base approach) because even though a wrong base may take longer to incorporate it may be associated with the primer for the same length of time as the correctly incorporated base.
In addition to false positive errors discussed above, false negatives can be a major problem in hybridisation based assays.
However, it is likely that false negatives will remain to some level.
However very often every probe will not bind to its complementary sequence and there may be gaps in the string of sites along the molecule.
It may be that the wrong strand has been captured by the array probes.
The greater problem will be when a non-functional duplicate of the sequence (e.g
pseudogene) becomes captured.
Although this kind of occurance can be detected when it is rare, it will be more difficult when it competes effectively with the functional sequence.
In some cases, despite stringency control, the probe may have bound but it may be a mismatch interaction.
It has been suggested that the bead like appearance is due to the fact the conditions used in denaturing the DNA actually cause the DNA chain to snap.
One problem is that often molecules that are stretched out on a surface undergo
light induced breakage.
However the longer the length the less easy it is to discriminate a single base difference by hybridisation.
It is recognised that hybridisation of
rare species is discriminated against under conventional
reaction conditions, whilst species that are rich in A-T base pairs are not able to hybridise as effectively as G-T rich sequences.
There is a concern that duplicated regions of the
genome may lead to errors, where the results of an
assay may be biased by DNA from a duplicated region.
The methods proposed by US
Genomics do not provide this and there are possibilities for incorrect positioning of sequences on a long range map.
If the
genome draft is solely used for this long range reconstruction then information of large scale duplications, amplifications, deletions, translocations etc may be lost.
Also it would take longer to complete the sequencing and length
sample preparation procedures would be required in advance of the sequencing run.
 Login to View More
Login to View More