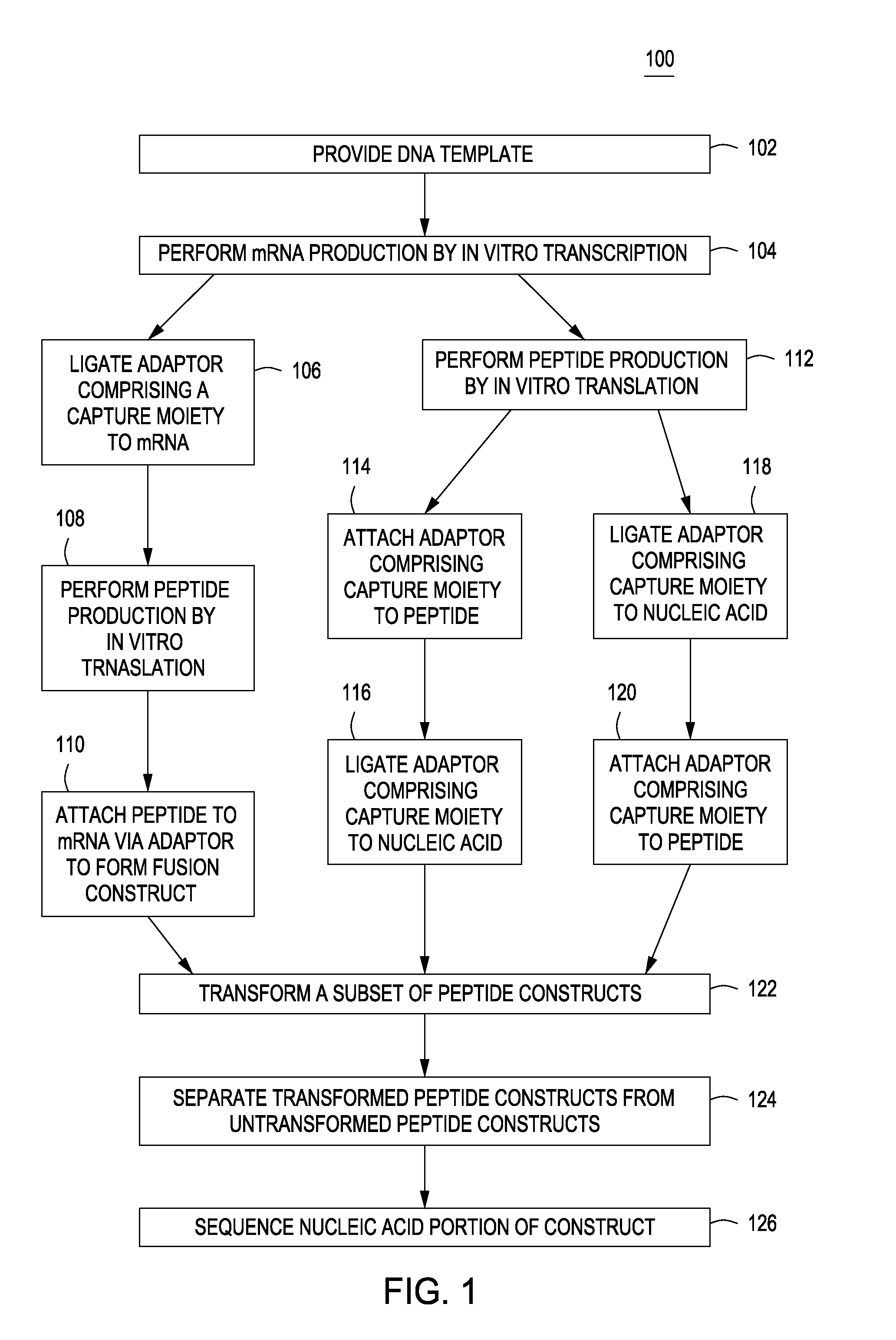
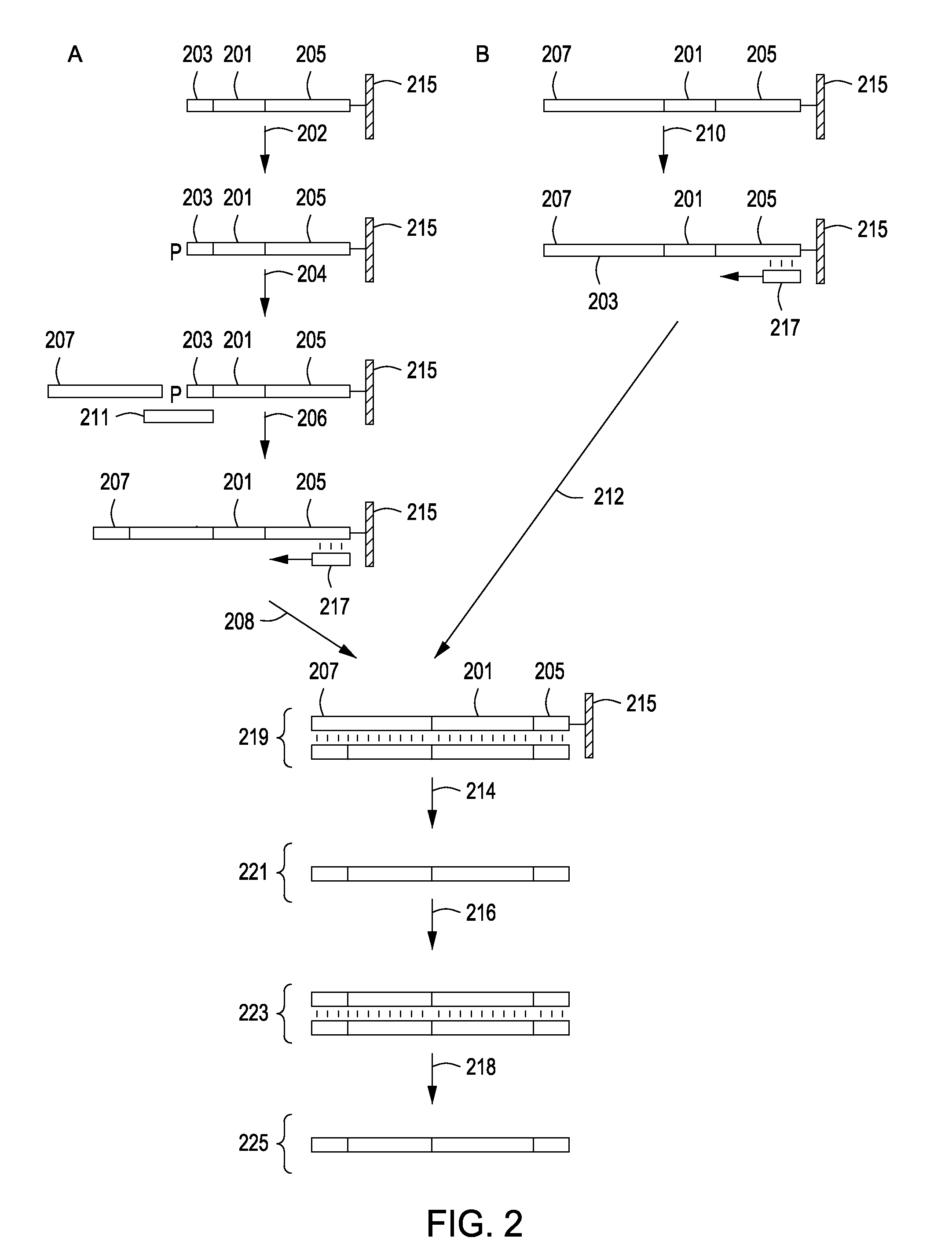
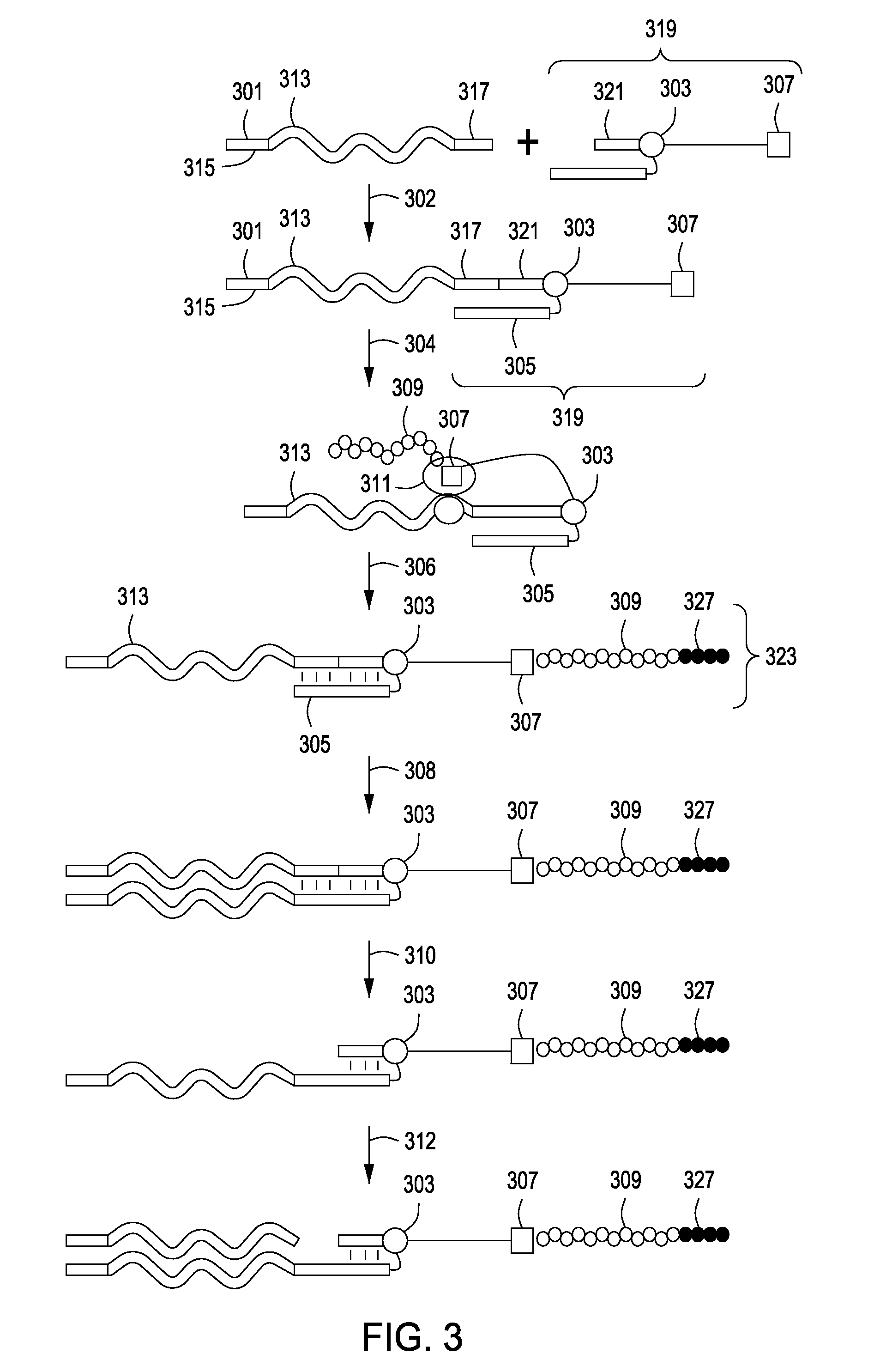
Peptide constructs and assay systems
a technology of constructs and assays, applied in the field of peptide constructs and assay systems, can solve the problems of inability to generate compact collections of protein sequences, inability to use highly specific and sensitive high-throughput methods for assaying proteins as a large collection, and inability to achieve large-scale proteomics studies. , to achieve the effect of facilitating separation of two populations, high through and high-throughput screening of peptides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Model System for Protease Substrate Generation and Screening
[0118]A model system was created to test the utility of the peptide constructs of the present invention in a protease assay. The model peptides used were GLVPRGSAG (SEQ ID NO:1), a target of commercially available thrombin (EMD Biosciences, San Diego, Calif.) and AGDDDDKAG (SEQ ID NO:2), a target of commercially available enterokinase (New England Biolabs, Ipswitch, Mass.). The protease recognition sites are shown in bold in the peptide sequences.
[0119]In addition to the peptide sequence of interest, peptide tags were chosen to be included in each peptide construct: Tag 1 at the N-terminus of all peptides, to be used in the protease substrate screening assay for peptide construct capture, and Tag 2 at the C-terminus as a tool to select for full-length peptides. FLAG (DYKDDDDK) (SEQ ID NO:3) and AU1 (DTYRYI) (SEQ ID NO:4) were analyzed for use as a C-terminal peptide Tag 2 for peptide construct purification. Inclusion of Tag...
example 2
Composition of a Set of Custom Peptide Protease Substrates
[0132]One peptide construct set was designed and used in an assay system to analyze protease cleavage sites within a viral genome. For this purpose, a 2,600-plex peptide construct set was created based on the methods developed and constructs produced in Example 1, though as noted in Example 1, the peptide construct sets could have been created using a Scheme II embodiment as an alternative to Scheme I. The construct set was designed to test the substrate specificity of NS3 (hepacivirin) protease of hepatitis C virus.
[0133]The HCV viral genome is a single-stranded, 9.5 kb long RNA molecule that is translated into a single polyprotein of about 3,000 amino acids. The NS3 (hepacivirin) protease of HCV is responsible for the cleavage at the NS3 / NS4A, NS4A / NS4B, NS4B / NS5A, and NS5A / NS5B sites of the polyprotein (Kwong, et al., Antiviral Res., 41(1):67-84 (1999)). It is essential for viral replication and the formation of infectious...
example 3
Peptide Constructs Generated from Fragmented Genomic DNA
[0143]The constructs and assay systems of the invention were also used to perform proteomic analysis of the genome of an organism, the yeast Saccharomyces cerevisiae. Genomic DNA from S. cerevisiae was sheared using adaptive focused acoustics technology for DNA shearing (Covaris), and then size selected to obtain random DNA fragments of approximately 100 base pairs. Custom DNA adaptors were attached to create constructs such as those illustrated in FIG. 7 at 701.
[0144]A genomic DNA library was created with approximately 100 bp fragment size from S. cerevisiae genomic DNA, and it was determined by sequencing that the initial DNA pool covered ˜93% of the genome. A set of peptide constructs was created from the genomic DNA library using the methods described in Example 1, although, again, a Scheme II ‘one-pot’ embodiment may have been employed as well. Upon sequence analysis following production of the peptide constructs, the pept...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com