Organic solvent resistant protease producing strain, gene of organic solvent resistant protease produced thereby and application of organic solvent resistant protease
A technology resistant to organic solvents and proteases, applied in applications, genetic engineering, plant genetic improvement, etc., can solve the problems of strong substrate selection specificity, low assay tolerance concentration, low enzyme yield, etc., to achieve high yield, The effect of high specific activity and huge application potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Use different concentrations of organic solvents such as cyclohexane and toluene as screening pressure to obtain organic solvent-resistant extremophiles from oily soil samples: use milk agar plate medium, the specific formula is (g / L): tryptone 5, yeast powder 3. Skim milk powder 25, agar 12. The screened organic solvent-resistant microorganisms were inoculated on milk agar plates, and the strains with high protease production were initially screened according to the ratio of the colony to the size of the transparent circle. This method screened 13 strains of extremophile resistant to organic solvents with high protease production.
[0027] In order to further test the solvent tolerance of the protease obtained from the preliminary screening, the protease production ability of the 13 strains and the organic solvent resistance of the protease produced were comprehensively tested. Inoculate the protease-producing bacterial strain obtained by preliminary screening into th...
Embodiment 2
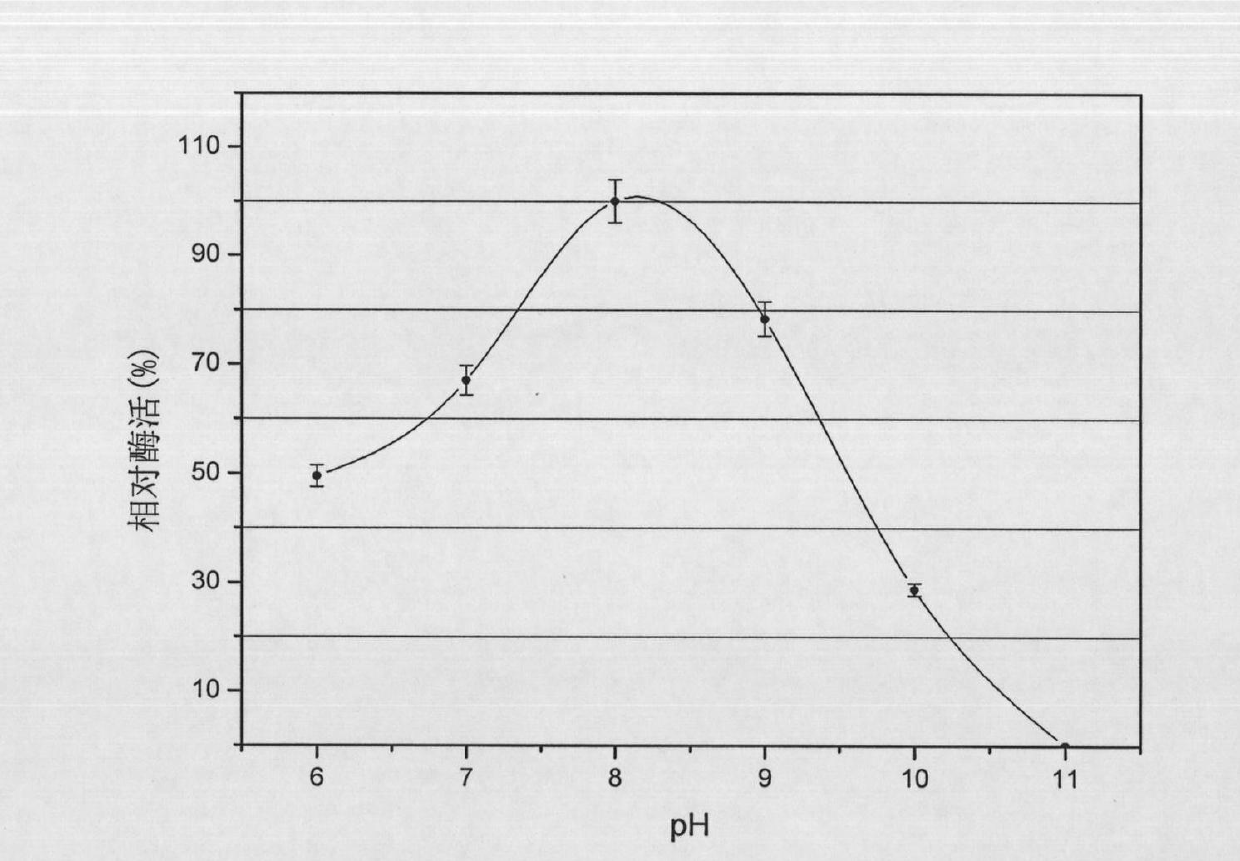
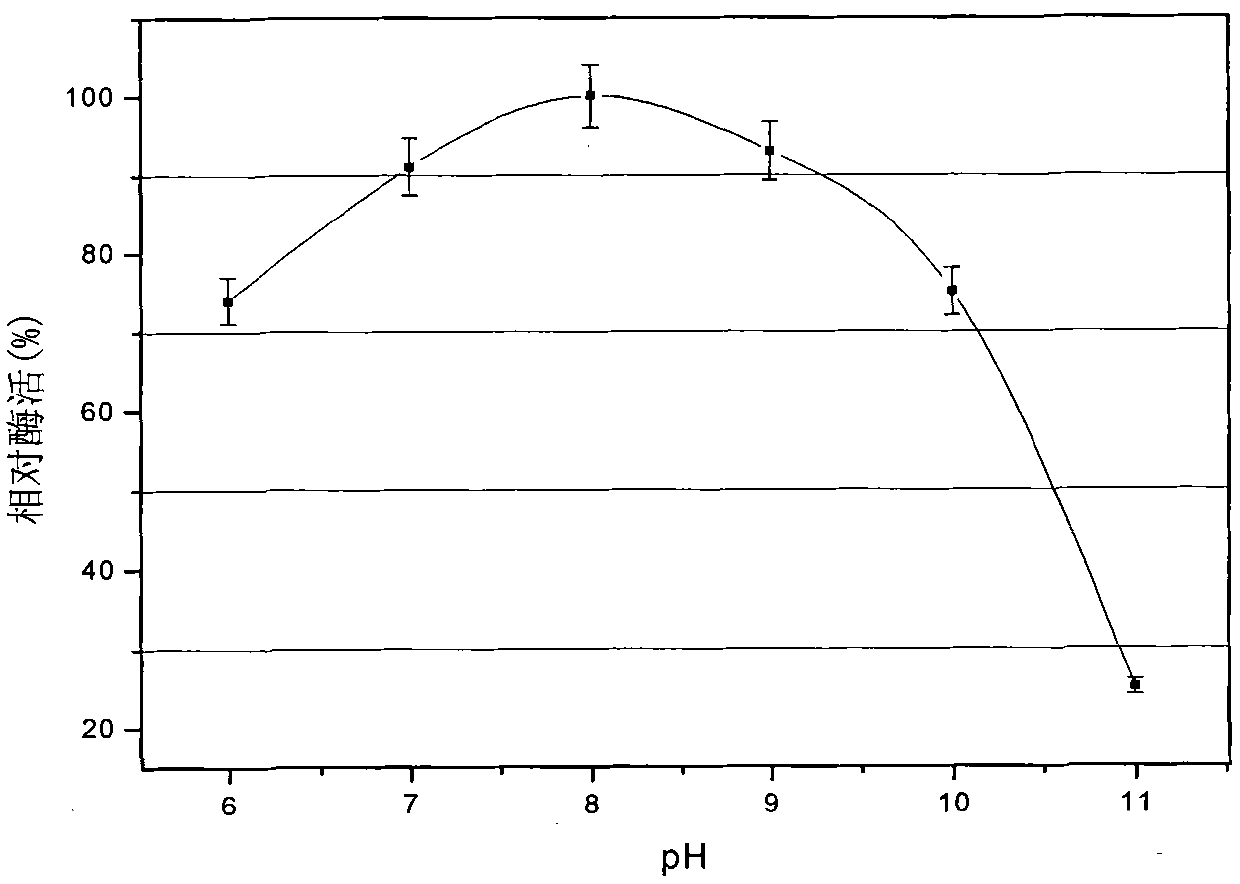
[0032] Biological properties of the strain WQ9-2: Gram staining showed that the strain was a Gram-positive strain with spores and grew under aerobic conditions. After growing in broth medium for 24 hours, the colony size and diameter are 3mm-5mm, the growth temperature range is 20°C-40°C, the optimum growth temperature is 35°C, the growth pH range is 6.0-11.0, and the optimum growth pH is 8.0 , its physiological and biochemical characteristics are negative in oxidase reaction, positive in the utilization of glucose, maltose, sucrose and D-fructose, and negative in the utilization of D-xylose and lactose.
[0033] Species identification of strain WQ9-2: The BIOLOG automatic bacterial identification instrument and 16SrDNA sequence analysis showed that the strain belonged to the genus Bacillus cereus, and it was named Bacillus cereus WQ9-2.
[0034] Study on fermentation conditions of Bacillus cereus WQ9-2 protease production: through single factor experiment and response surface...
Embodiment 3
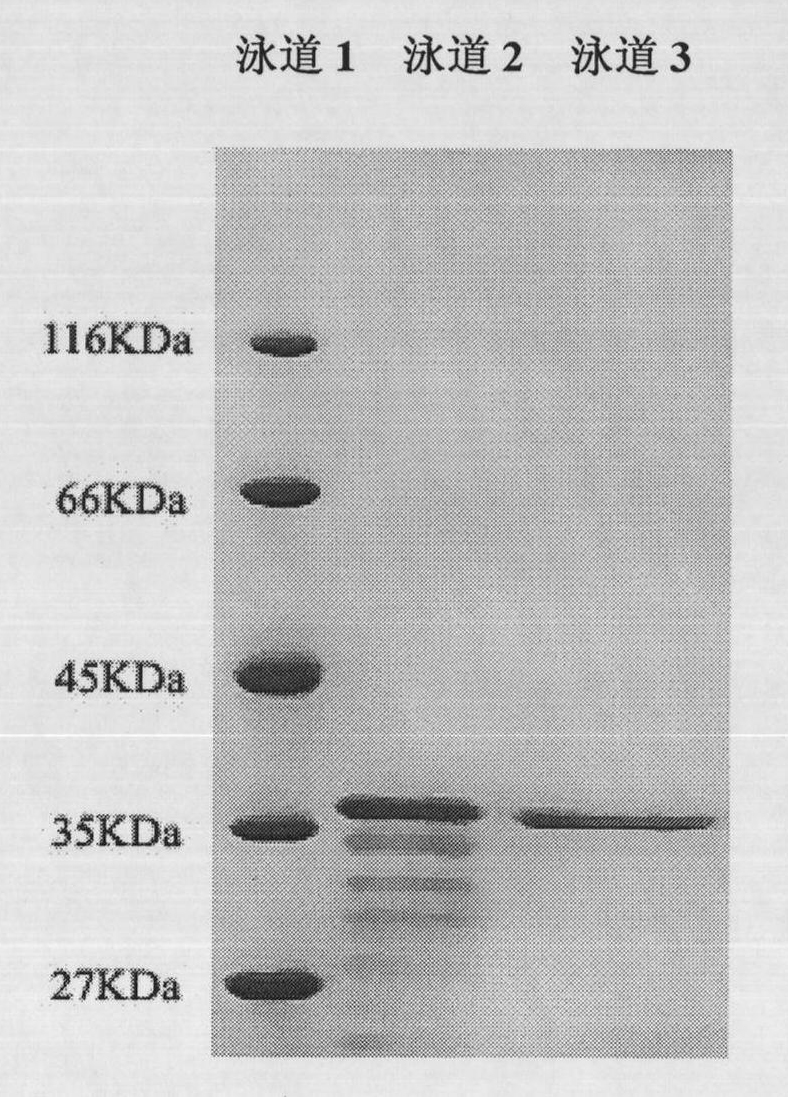
[0037] After culturing Bacillus cereus WQ9-2 in the enzyme-producing medium for 48 hours, the fermentation broth was centrifuged at 10,000 rpm and 4°C for 10 minutes, and the supernatant was taken as the crude enzyme solution; the crude enzyme solution was placed in an ice bath and collected by ethanol precipitation 45%-75% (v / v) part of the precipitated protein was then dissolved with 0.05M Tris-HCl buffer solution of pH 8.5, and centrifuged at 10,000 rpm at 4°C for 10 min to remove insoluble miscellaneous proteins. The enzyme solution obtained from the above treatment was purified with a DEAE-Sepharose FF ion-exchange chromatography column, and eluted with 0.05M Tris-HCl (pH 8.5) buffer solution to collect protease activity peaks. By SDS-PAGE electrophoresis ( figure 1 ), found that the two-step purified organic solvent-resistant protease (named as organic solvent-resistant WQ9-2 protease) had reached electrophoretic purity, and the molecular weight of the protease subunit w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com