A method for improving the thermal stability of α-l-rhamnosidase r-rha1
A technology of rhamnosidase and thermal stability, applied in the fields of genetic engineering and enzyme engineering, to achieve the effect of excellent enzymatic properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Acquisition of α-L-rhamnosidase gene
[0029] Escherichia coli DH5α containing WT (pPIC9K-rha) plasmid was inoculated in 30 mL LB liquid medium containing 1 mg / mL ampicillin resistance and cultured at 37 ° C for 16 h.
[0030] The WT (pPIC9K-rha) plasmid was extracted using the plasmid mini-extraction kit (Takara Company) according to the instructions. EcoRI and BlnI double enzyme digestion was used for verification. After verifying that the result is correct, it is used as a template for error-prone PCR.
Embodiment 2
[0031] Example 2: Construction and screening of α-L-rhamnosidase mutant library
[0032] The WT gene was subjected to error-prone PCR to establish a mutant library, and the enzyme was induced and expressed in a 96-microwell plate. After screening, a mutant with significantly improved thermal stability was obtained.
[0033] Using the WT (KC750908.1) gene, design a pair of specific primers:
[0034] Upstream primer Q9KF (SEQ ID NO.3): 5'-CCG GAATTC GTACCCTACGAGGAGTACATTTCTAG-3′,
[0035] Downstream primer Q9KR (SEQ ID NO.4): 5'-CGC CCTAGG TTACACATTCAACCGCCATTTC-3';
[0036] The PCR reaction conditions were: pre-denaturation at 94°C for 4 min, denaturation at 94°C for 1 min, annealing at 50°C for 1 min, extension at 72°C for 2.5 min, and after 35 cycles, extension at 72°C for 7 min. PCR products were purified using Takara PCR Product Column Recovery Kit.
[0037] Recombinant expression plasmids were constructed by restriction cloning, that is, the error-prone PCR products...
Embodiment 3
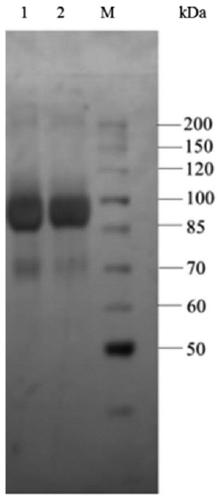
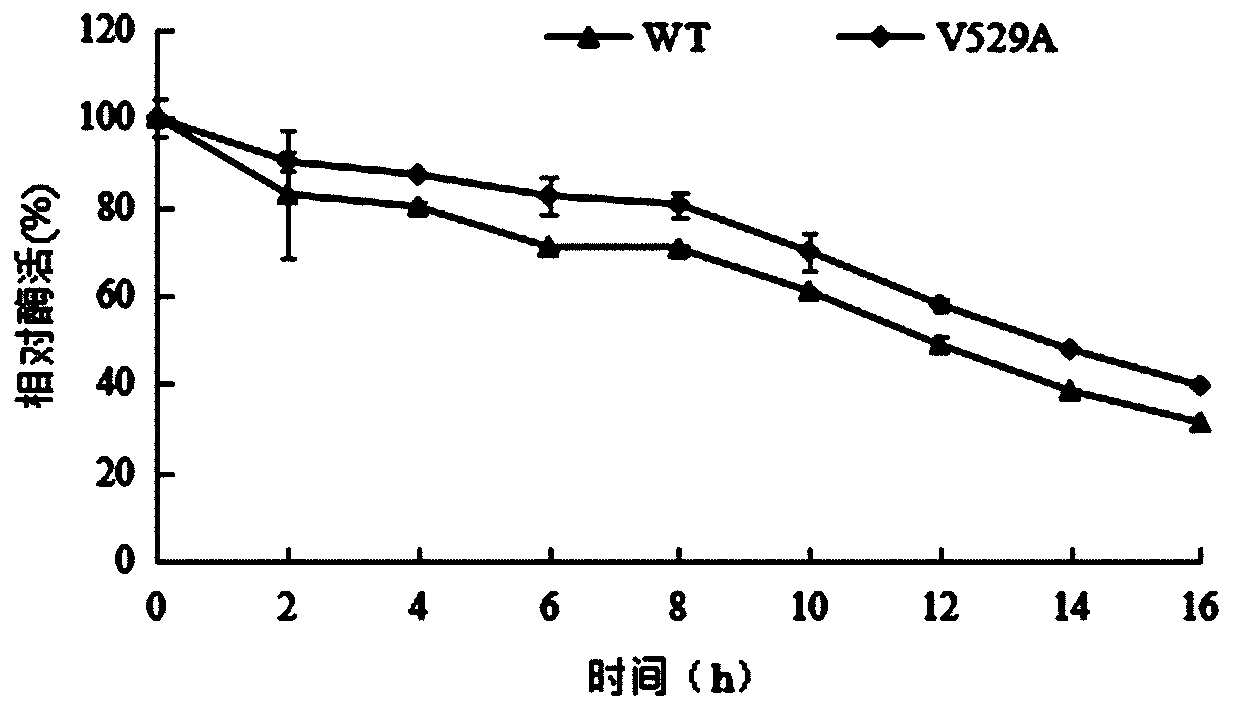
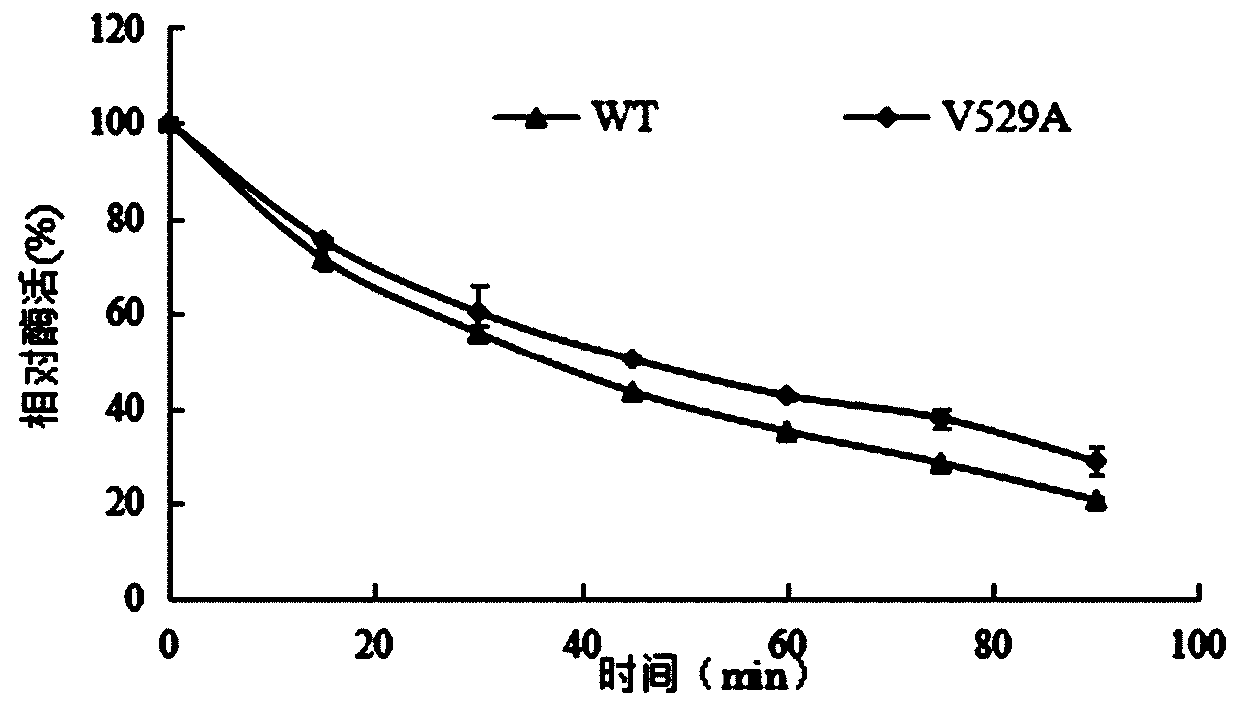
[0039] Example 3: Expression and purification of α-L-rhamnosidase WT and V529A using recombinant expression strains
[0040] Inoculate the strains with 1% inoculum in 50mLYPD liquid medium for strain activation, shake culture at 30°C for 16h; min, after culturing for 16 hours, measure and determine its OD 600Reach the range of 3.0-5.0; centrifuge for 10 minutes to collect all the bacteria, discard the supernatant, transfer all the bacteria to 100mL BMMY medium, 30°C, culture for 7 days, add 0.5% to the medium every 24h during the culture period Sterile methanol solution; after cultivation, the supernatant collected by centrifugation is the enzyme solution.
[0041] The α-L-rhamnosidase WT and V529A crude enzyme solutions were collected, concentrated by ultrafiltration through a 30kDa membrane, and set aside. At a flow rate of 0.5mL / min, use 20mmol / L of C 6 h 8 o 7 -Na 2 HPO 4 buffer and 0.15mol / L sodium chloride solution on Sephacryl TM Equilibrate S-200HR, load the co...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com