Chlamydomonas reinhardtii hemolytic phosphatidic acid acyltransferase and its gene and application
An acyltransferase, hemolytic technology, applied in the field of molecular biology and biology, can solve problems such as unclear LPAAT, and achieve the effect of low viscosity and low ignition point
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1 gene cloning and sequence feature analysis
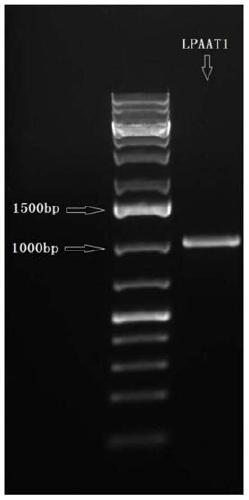
[0048] The CrLPAAT1 gene provided by the present invention is based on the Cre09.g398289.t1.1 gene published by the JGI Chlamydomonas reinhardtii genome database (http: / / phytozome.jgi.doe.gov / pz / portal.html) as a reference gene, and the design is specific Primer (forward primer, as shown in SEQ ID No.3, 5'-ATGGCGCGTAAAAGCAGTTTGGCTC-3', reverse primer, as shown in SEQ ID No.4, 5'-CTGCTCATCCGGCGCCATCTCTGT-3'), using Chlamydomonas reinhardtii cDNA The library (the construction of the cDNA library is prepared and stored according to the "Molecular Cloning Experiment Guide" by conventional methods) is used as a template, and the cDNA sequence is obtained by PCR amplification, such as figure 1 shown. The PCR reaction system was: 2 μl template DNA, 10 μl 2×GoTaq DNA polymerasemix (purchased from Promega (Beijing) Biotechnology Co., Ltd.), 1 μl forward primer (10 μM), 1 μl reverse primer (10 μM), and finally supplemente...
Embodiment 2
[0051] Example 2 Function Verification
[0052] Using Chlamydomonas reinhardtii cDNA as a template, primers (forward primer, shown in SEQ ID No.5, 5'-tgacggatccATGGCGCGTAAAAGCAGTTTGGCTC-3', reverse primer, As shown in SEQ ID No.6, 5'-actgaagcttTCACTGCTCATCCGGCGCCA TCTCTGT-3') and high-fidelity polymerase pfu were amplified by PCR to obtain 5' and 3' open reading frame DNA fragments containing corresponding restriction sites. T4 DNA ligase was used to connect the fragment with the double-digested expression vector pQE30 to obtain a recombinant plasmid. The recombinant plasmid was transformed into LPAAT-deficient temperature-sensitive Escherichia coli SM2-1 (plsC-) (SM2-1 was obtained from Yale University Escherichia Coli Resource Center CGSC) competent cells by heat shock method. Compared with the control group (containing the pQE30 empty vector), the SM2-1 strain transformed with the CrLPAAT1 gene could survive at 42°C after the recombinant protein was induced by IPTG, while ...
Embodiment 3
[0054] Example 3 In vitro enzyme activity test
[0055] In the present invention, the SM2-1 Escherichia coli transformed with the CrLPAAT1 gene and the SM2-1 Escherichia coli transformed with the pQE30 empty plasmid were cultured at 30° C. and 0.5 mM IPTG was added to induce protein expression for 6 hours, and the cells were collected by centrifugation. The membrane fraction was extracted by mechanical crushing and gradient centrifugation and used as a crude enzyme for in vitro enzyme activity experiments. In the experiment, a variety of acyl-CoAs with different carbon chain lengths and degrees of saturation were selected as acyl donors, and the optimal substrate was determined by observing the accumulation of the product phosphatidic acid (PA). The conditions of the enzyme reaction are: 250 μM lysophosphatidic acid (LPA), 200 μM acyl-CoA (Acyl-CoA), 1 mM MgCl in 200 μl 100 mM potassium phosphate buffer (pH 7.4) 2 with 20 μg crude enzyme. After the crude enzyme was added, th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com