Product and method for detecting clinic common pathogenic bacteria in blood culture bottle
A technology for pathogenic bacteria and products, applied in the field of microbial detection, can solve the problems of high cost, inability to meet clinical large-scale bacterial detection, complicated operation, etc., and achieve the effect of reducing the cost of consumables
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1: Primer Design and Synthesis
[0053] For Klebsiella pneumoniae khe gene, Acinetobacter baumannii OXA gene, Streptococcus pneumoniae LytA gene, Enterococcus faecium and Enterococcus faecalis ddl gene, Escherichia coli phoA3 gene, Staphylococcus aureus nuc gene, Staphylococcus epidermidis Design specific primers for 2312 gene, Pseudomonas aeruginosa ecfX gene and Proteus ure gene. The specific primer sequences are shown in Table 1 above.
[0054] Related primers were synthesized at Sangon Bioengineering (Shanghai) Co., Ltd.
Embodiment 2
[0055] Embodiment 2: sample DNA extraction
[0056] A total of 150 samples of clinical blood culture bottles were collected, and the bacterial genomic DNA in the blood culture bottles was extracted by the guanidine hydrochloride-benzyl alcohol extraction method.
[0057] (1) Aseptically take 100uL of the culture from the positive blood culture bottle and put it into a 1.5mL EP tube, then add guanidine hydrochloride buffer and vortex for 3min.
[0058] (2) Add 100uL double-distilled water and 400uL benzyl alcohol in turn, vortex for 1min, and then centrifuge for 5min.
[0059] (3) Transfer the upper aqueous phase to a new EP tube, add 40 uL of sodium acetate buffer solution and 440 uL of isopropanol in sequence, and then centrifuge at high speed for 15 min.
[0060] (4) Discard the upper layer liquid, add 1mL of 70% ethanol to wash, then centrifuge at high speed for 5min, then discard the upper layer of ethanol, after natural drying, add 100uL double distilled water to dissolv...
example 3
[0061] Example 3: Biological Experiment
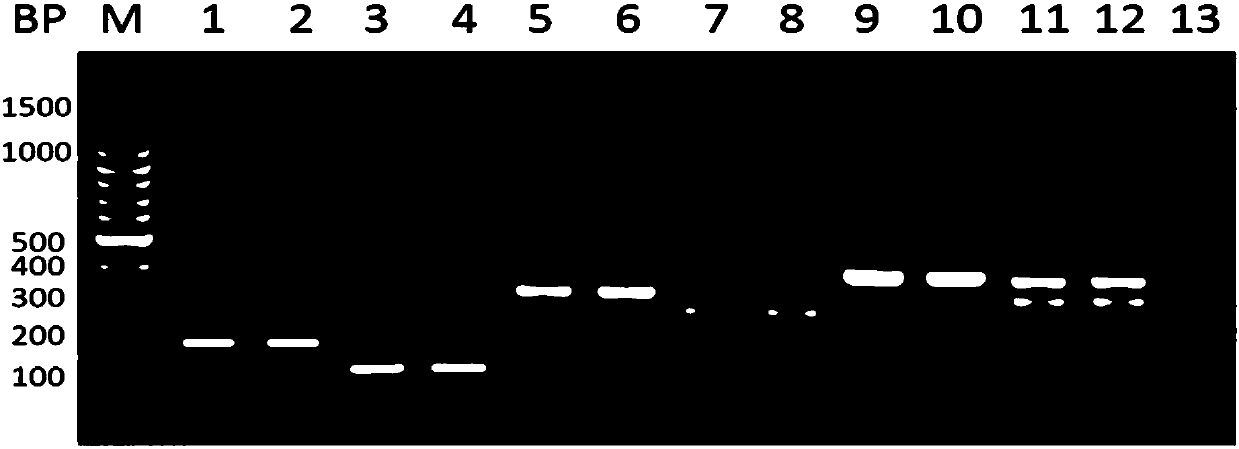
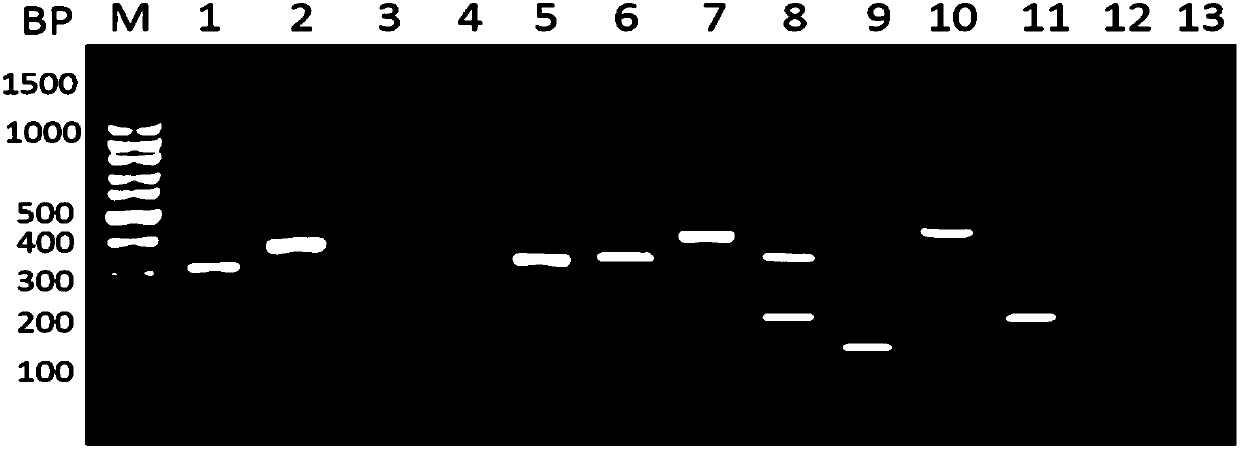
[0062] 1. Multiplex PCR
[0063] System: 10 μL total volume
[0064] Hot start enzyme 0.05 μL;
[0065] 1 μL bacterial DNA template;
[0066] The upstream and downstream primers for each type of bacteria were mixed at a concentration of 1 μM, and 1 μL was taken;
[0067] Sterile ultrapure water 2.95μL;
[0068] PCR reaction buffer 5mL
[0069] (10 kinds of bacterial primers are divided into two groups, added in different reaction gates, each group has 5 different bacteria).
[0070] Reaction parameters:
[0071] Pre-denaturation at 95°C for 5 minutes;
[0072] Denaturation at 95°C for 30s;
[0073] Anneal at 59°C for 90s;
[0074] Extend at 72°C for 60s;
[0075] A total of 35 cycles were performed, and the cycle was extended at 72° C. for 10 min.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com