Method for constructing exoglucanase Cel6A and Cel7A in heterologous expression in pichia pastoris
A technology of exoglucanase and Pichia pastoris, which is applied in the field of genetic engineering, can solve the problems of low efficiency and lack of enzymatic hydrolysis of cellulase system, and achieve the potential of improving industrial production, increasing the success rate and reducing errors. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment example 1
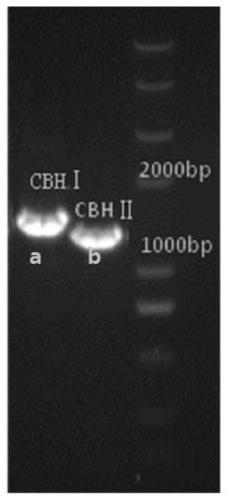
[0028] Example 1 Cloning of the target gene
[0029] 1. Take the Trichoderma reesei QM9414 spore suspension stored in a glycerol tube out of the -80°C refrigerator and place it in ice (or 4°C refrigerator) to thaw until the spore suspension is completely thawed. The spore suspension was shaken and mixed evenly, a small amount of bacterial liquid was dipped in the inoculation ring, and streaked on the PDA medium plate for inoculation. After sealing the plate with parafilm, it was cultured in a 28°C incubator. Culture for about 5-7 days until the surface of the medium is covered with green spores. Remove the plate from the incubator and place it in a 4°C refrigerator for later use.
[0030] 2. Inoculate the fresh spores on the cellulase solid induction culture, cover the surface with sterilized cellophane, and let it stand for culture until the mycelium grows out. The hyphae were scraped from the cellophane and treated with a liquid nitrogen freezer grinder. The UNIQ-10 colum...
Embodiment example 2
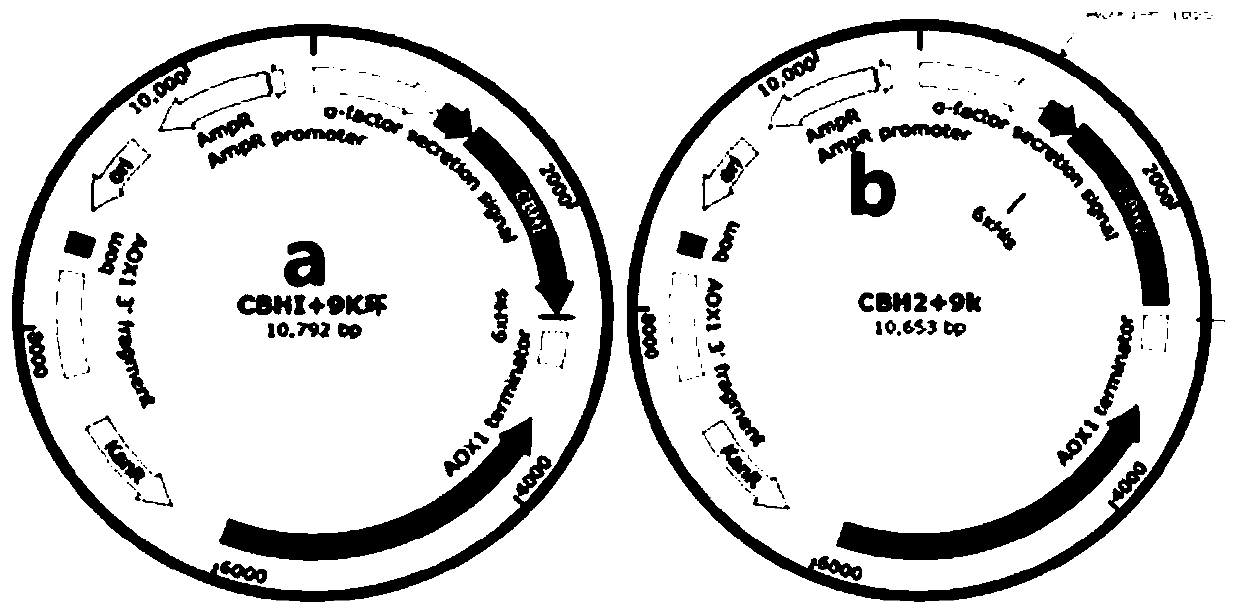
[0032] Example 2 Construction of Ppic9k-Cel6A, Cel7A expression vector
[0033] 1. First put the Buffer in ice, and after it is fully melted and mixed evenly, prepare a double-enzyme digestion reaction system. After the preparation is completed, mix it evenly, place it in a 37°C water bath, and perform enzyme digestion for 30 minutes. After the end of the digestion, use a purification and recovery kit to recover the digestion product to remove the residual enzyme and oligonucleotide in the digestion product, so as not to affect the subsequent connection of the plasmid and the target gene.
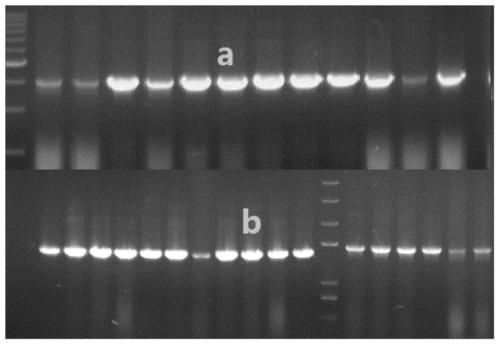
[0034] 2. Using the 5'-end EcoRI as the restriction site and the 3'-end Not I as the restriction site, insert it into the downstream of the promoter AOX1 of the Ppic9k plasmid vector to construct the expression vector Ppic9k-Cel6A, Cel7A, see the expression vector map image 3 . Colony PCR validation see figure 2 . The expression vector was verified by single enzyme digestion to confirm...
Embodiment example 3
[0035] Example 3 Linearization of expression vectors Ppic9k-Cel6A, Cel7A and electrotransformation
[0036] 1. Take SaII as the linearization site of the Ppic9k-Cel6A and Cel7A expression vectors to realize the linearization of the expression vectors, and verify the linearized plasmids by nucleic acid electrophoresis, see Figure 4 . Transformed into Pichia pastoris GS115 by electroporation, and plated to obtain transformants.
[0037] 2. Use SDS pyrolysis method to crudely extract the genome of the recombinant yeast transformant obtained from 1, and verify its correctness and phenotype by PCR. See the genome PCR nucleic acid map. Figure 5 .Use antibiotic concentration gradient to screen multi-copy transformants, and screen out the colonies that can grow normally on high-concentration resistant plates.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com