Streptomyces-sourced broad-spectrum polysaccharide degrading enzyme rAly16-1 as well as coding gene and application thereof
A technology of polysaccharide degrading enzyme and coding gene, which is applied in the field of broad-spectrum polysaccharide degrading enzyme rAly16-1 and its coding gene and application, can solve the problems of precise application and directional molecular enzymatic transformation, lack of mechanism and cognition, etc. Achieve the effect of stable physical and chemical properties and high activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0114] Extraction of Genomic DNA from Streptomyces sp. CB16 Strain
[0115] Streptomyces sp. CB16 strain was inoculated into TSB liquid medium, and cultured with shaking at 28°C and 200 rpm until the absorbance value at 600 nm (OD 600 ) was 1.21; take 20 mL of the cultured bacteria, centrifuge at 28°C and 12,000×g (g, the earth’s gravitational constant) for 20 minutes, and collect the bacterial sediment.
[0116] The above TBS liquid medium is composed of: tryptone 17g / L, plant peptone 3g / L, sodium chloride 5g / L, potassium dihydrogen phosphate 2.5g / L, glucose 2.5g / L, pH 7.2.
[0117] Add 12.0 mL of lysozyme buffer solution to the above cell precipitation to obtain about 14.0 mL of bacterial liquid, add 560 μL of lysozyme with a concentration of 20 mg / mL respectively, and the final concentration is about 800 μg / mL; place in an ice-water bath for 1.0 h , and then transferred to a 37°C water bath, warmed for 2h until the reaction system was viscous; added 0.82mL of sodium hexade...
Embodiment 2
[0119] Genome Scanning and Sequence Analysis of Streptomyces sp. CB16 Strain
[0120] The scanning and sequencing of the genomic DNA prepared in Example 1 was carried out by pyrosequencing technology, which was completed by Shanghai Meiji Biotechnology Company. The DNA sequencing results were analyzed using the online software of the NCBI (National Center for Biotechnology Informationh, ttp: / / www.ncbi.nlm.nih.gov / ) website. The analysis software used on the NCBI website is Open Reading Frame Finder (ORF Finder, http: / / www.ncbi.nlm.nih.gov / gorf / gorf.html) and Basic Local Alignment Search Tool (BLAST, http: / / blast.ncbi.nlm.nih.gov / Blast.cgi).
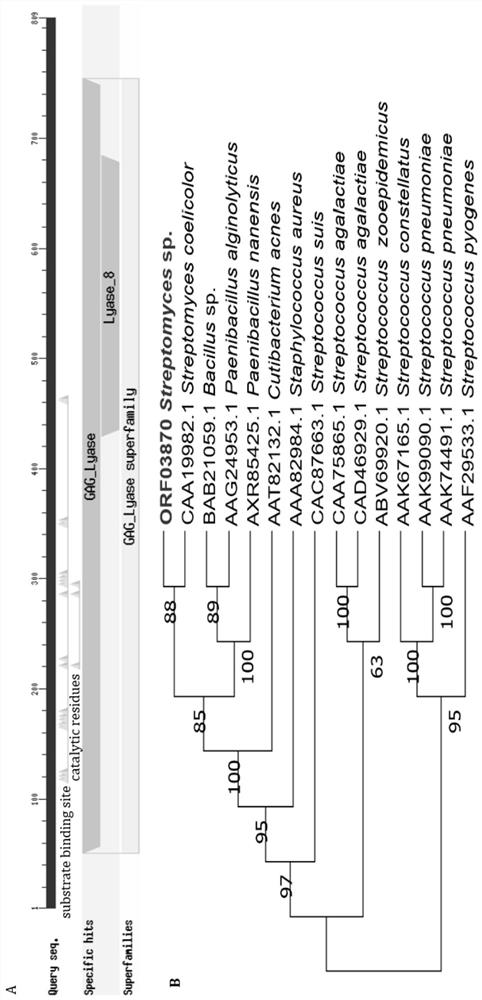
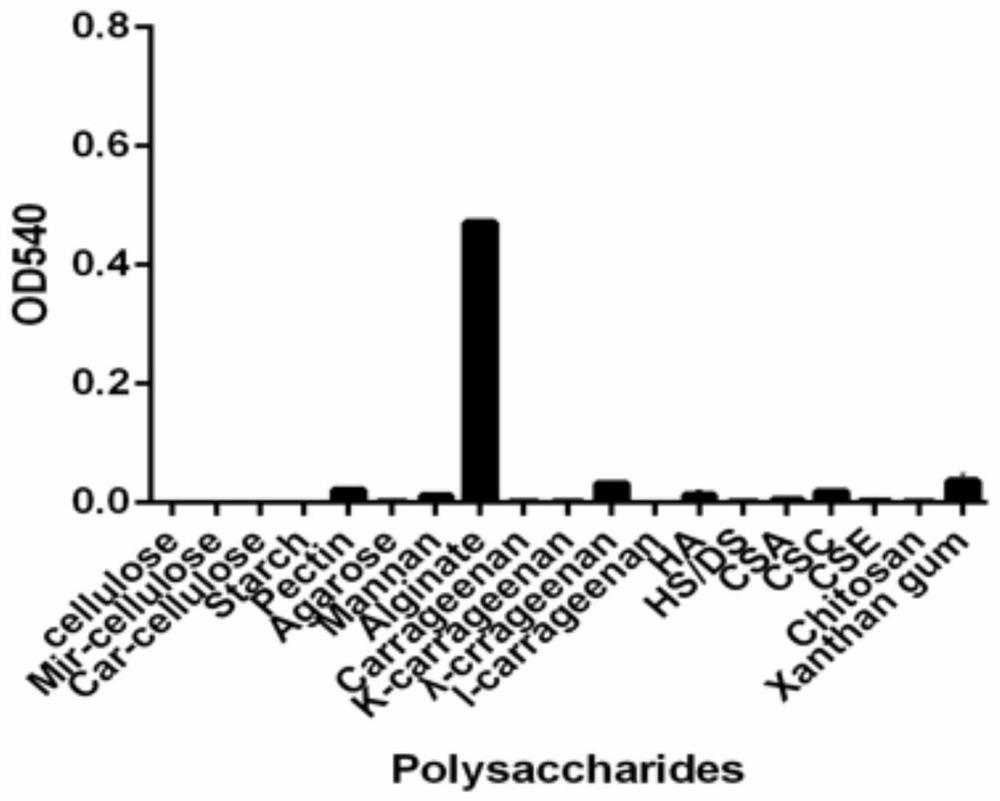
[0121] The results analyzed by the above biological software showed that Streptomyces sp. (Streptomyces sp.) CB16 strain genomic DNA carried a candidate polysaccharide degrading enzyme gene orf03870, the nucleotide sequence of which was shown in SEQ ID NO. There are 809 amino acids in the enzyme rAly16-1, and the 1-38 amino acids at th...
Embodiment 3
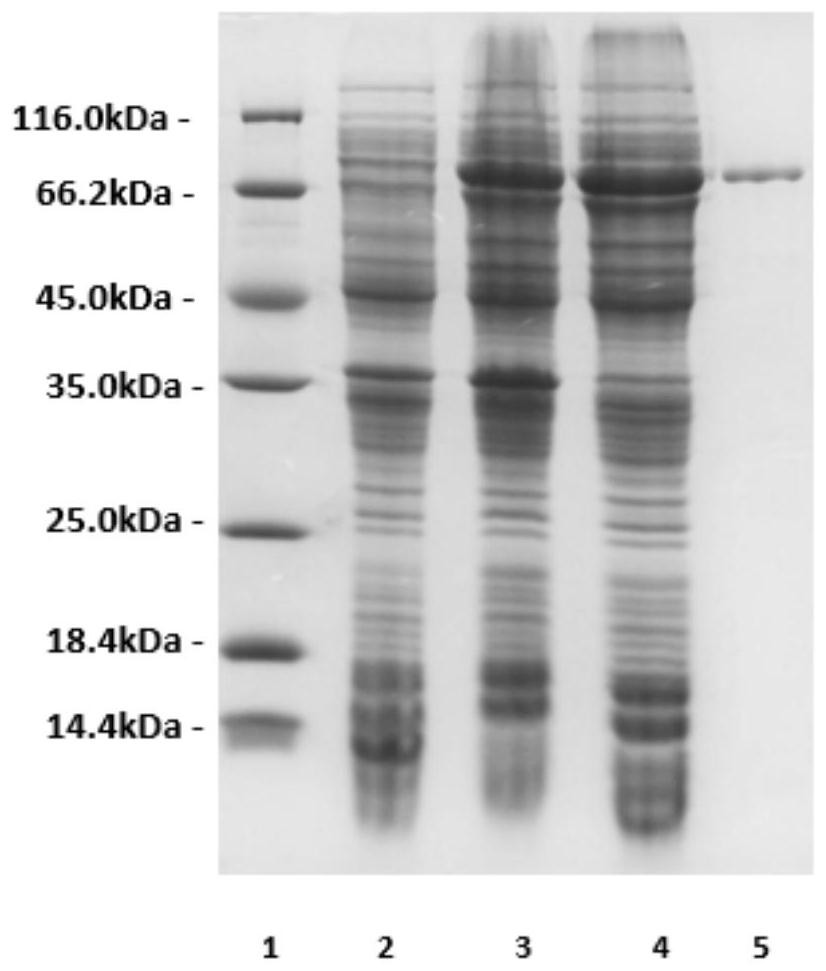
[0123] Recombinant expression of gene orf03870 in Escherichia coli BL21(DE3) strain
[0124] Using the genomic DNA prepared in Example 1 as a template, PCR amplification was performed. The primer sequences are as follows:
[0125] Forward primer orf03870-F: 5'-g catatg TCCGGCACGGCACGGACCG-3' (Nde I) SEQ ID NO.4;
[0126] Reverse primer orf03870-R: 5'-g tctaga GCTCCTCTTCAGGGTGAGGCC-3' (Xba I) SEQ ID NO. 5;
[0127] The underlined mark in the forward primer orf03870-F is the restriction endonuclease Nde I site, and the underlined mark in the reverse primer orf03870-R is the restriction endonuclease Xba I site. The high-fidelity DNA polymerase PrimeSTAR HSDNA Polymer used was purchased from China Dalian Biotech Co., Ltd., and the PCR reaction reagents used were operated according to the product instructions provided by the company.
[0128] PCR reaction conditions: pre-denaturation at 94°C for 5 minutes; denaturation at 98°C for 30 seconds; annealing at 65°C for 30 seconds...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com