Screening and application of enterococcus faecium G12
A technology of Enterococcus faecium and G12, applied in the field of microbial additives for aquatic products, can solve the problems of increased overall drug resistance and unfavorable treatment of animal diseases, and achieve the effect of sustainable development, significant probiotics, and high resistance to stress.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1: Screening, isolation and identification of strains
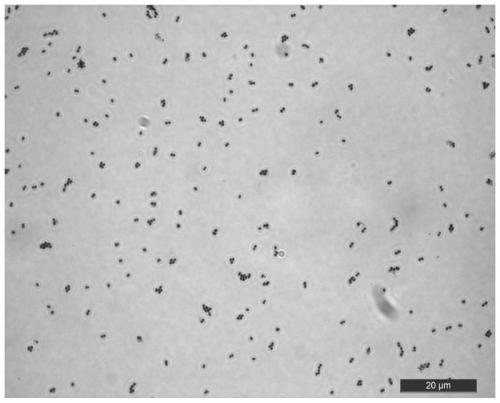
[0028] Screening and isolation: A strain of Enterococcus faecium was screened from the hepatopancreas of shellfish, especially the hepatopancreas of Philippine clams. The specific operation of the screening is as follows: Dissect the body surface of Philippine clams after disinfection with 75% medical alcohol, take the hepatopancreas, then add 1mL pre-cooled sterile saline, fully homogenate, centrifuge, and take the supernatant 10 times series dilution. Take stock solution, 10 -1 、10 -2 Each 100uL sample of the three dilutions was spread on the MRS plate, and three parallels were set for each dilution gradient, and incubated at a constant temperature of 37°C for 24h. Pick single colonies and purify bacteria by multiple streaking. The purified strains were made into bacterial suspension with 40% glycerol and MRS liquid medium 1:1, and stored at -80°C for future use. The growth state of the strain on MRS...
Embodiment 2
[0035] Embodiment 2: antibacterial test
[0036] Aeromonas vernerii, Pseudomonas aeruginosa and Vibrio vulnificus (the above strains were all provided by the Aquatic Animal Disease and Immunology Laboratory of Fisheries College of Tianjin Agricultural University) were used as indicator bacteria. Method With reference to the literature (Evidence for the competitive exclusion of Aeromonas salmonicida from fish with a tressinducible furunculosis by a fluorescent pseudomonad.Smith P and Dabey S, "Journal of Fish Diseases", No. 16, 1993), the activated indicator bacteria and the tested The strains were cultured in LB liquid medium for 24 h and diluted to 1×10 with saline 7 cfu / mL, take 0.2mL of the indicator strain and smear it on fresh LB solid medium, then take 2μL of the strain to be tested and inoculate it on the above plate, and observe whether there is an obvious inhibition zone around the inoculation area within 24 hours.
[0037] Re-screening was carried out by the Oxford ...
Embodiment 3
[0040] Example 3: Security Test
[0041] Hemolysis test: Streak culture of the strain to be tested on LB solid medium for 24 hours, scrape a single colony with an inoculation loop and inoculate it on a blood plate, incubate at 37°C for 24 hours, observe whether a transparent circle is formed around the colony, and judge the production of hemolysin . The experimental results showed that no hemolysis was observed on the blood plate, indicating that the strain did not produce hemolysin and had no potential pathogenicity.
[0042] Animal test: refer to the literature, select the immersion test on juvenile Penaeus monodon, select 1×10 7 , 1×10 5 Soak in cfu / mL bacterial liquid concentration, select Vibrio vulnificus bacterial liquid for positive control, and select normal saline for negative control group. After four hours of immersion in the bacterial suspension, the juvenile shrimp were transferred to aerated water. Each treatment group was performed in triplicate, observed f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com