Buffer solution for enhancing activity of TET enzyme
A technology of buffer solution and reaction buffer solution, applied in the field of biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
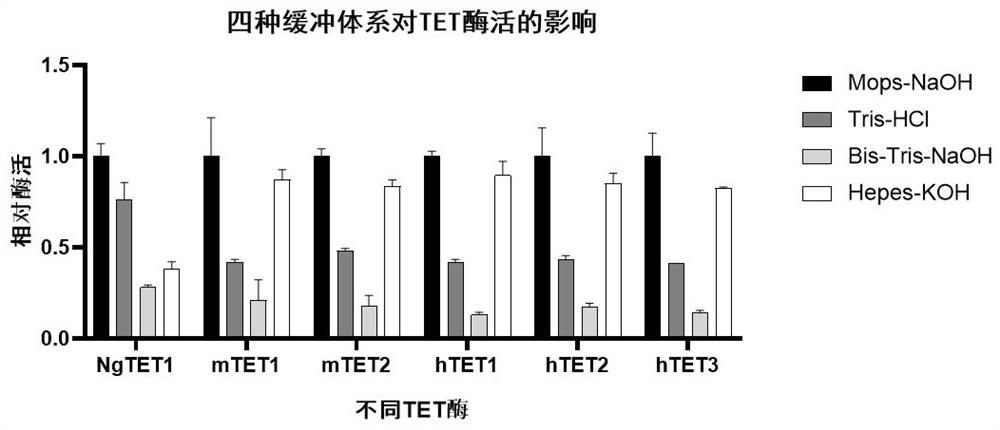
[0027] Example 1: Effects of four buffer systems on TET enzyme activity.
[0028] In this example, we verified the 3-(N-morpholino)propanesulfonic acid sodium salt-sodium hydroxide buffer system (Mops-NaOH), tris-hydrochloric acid buffer system (Tris-HCl) , bis-hydroxyethyltrismethylolmethylamine-sodium hydroxide buffer system (Bis-Tris-NaOH) or 4-hydroxyethylpiperazine sulfuric acid-potassium hydroxide buffer system (Hepes-KOH) four reactions The impact of the system on each TET enzyme, the specific implementation is as follows:
[0029] TET enzyme reaction buffer includes 10-100 mM buffer system (pH 7.5), 10-300 mM sodium chloride, 0.1-10 mM ascorbic acid (L-AA), 0.1-10 mM s adenosine triphosphate (ATP), 0.1-10 mM α - Ketoglutarate (α-KG), 30-300 uM ferrous ammonium sulfate, 0.1-5 mM dithiothreitol.
[0030] Epigentek's Epigenase 5mC-hydroxylase TET activity / inhibition assay kit (fluorescence method) was used to measure NgTET1, mTET1, mTET2, hTET1, hTET2, and hTET3 accordi...
Embodiment 2
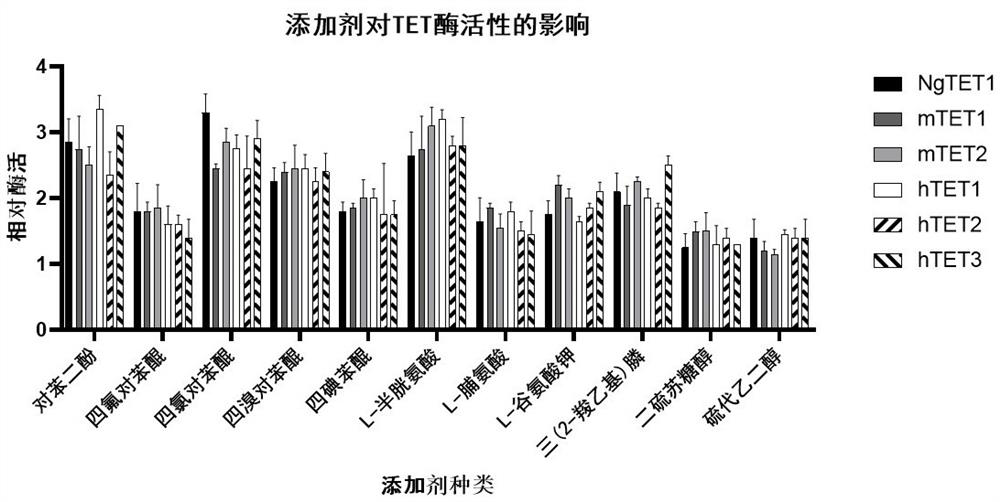
[0031] Example 2: Effect of small molecule additives on TET enzyme activity.
[0032] In this example, we verified the effects of different small molecule additives on TET enzyme activity. Small molecule additives are 1 mM hydroquinone, 1 mM tetrafluoro-p-benzoquinone, 1 mM tetrachloro-p-benzoquinone, 1 mM tetrabromo-p-benzoquinone, 1 mM tetraiodobenzoquinone, 1 mM L-cysteine , 1 mM L-proline, 1 mM potassium L-glutamate, 1 mM tris(2-carboxyethyl)phosphine, 1 mM dithiothreitol, 1 mM thioethylene glycol. Using Epigentek's Epigenase 5mC-hydroxylase TET activity / inhibition assay kit (fluorescence method), add the above-mentioned small molecule additives respectively, and measure the enzyme activity of TET according to the procedure in the manual. The enzyme activity assay results were as figure 2 , phenol and quinone compounds (hydroquinone, tetrafluoro-p-benzoquinone, tetrachloro-p-benzoquinone, tetrabromo-p-benzoquinone and tetraiodobenzoquinone), amino acid analogs (L-cystei...
Embodiment 3
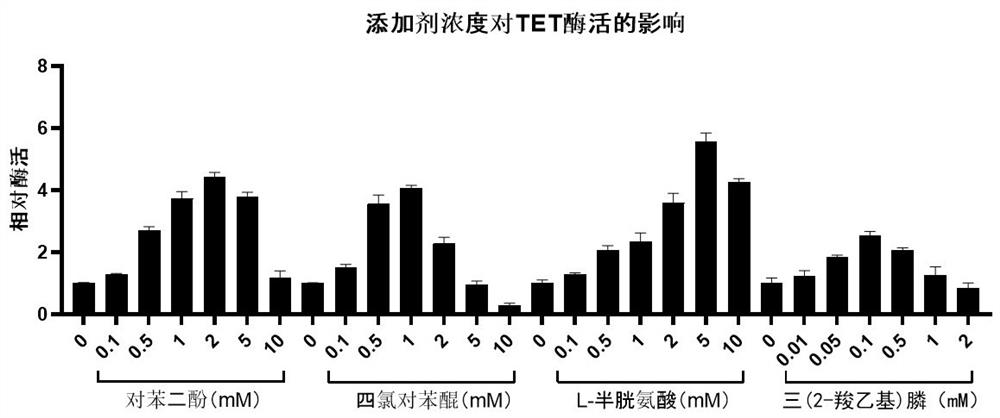
[0033] Example 3: Effects of concentrations of hydroquinone, chloro-p-benzoquinone, L-cysteine and tris(2-carboxyethyl)phosphine on TET enzyme activity.
[0034] In this example, we verified the effects of hydroquinone, chloro-p-benzoquinone, L-cysteine and tris (2-carboxyethyl) phosphine on the enzyme activity of NgTET1 in the TET enzyme reaction buffer. Refer to Examples 1 and 2 for the flow process. The result is as image 3 As indicated, 1-5 mM hydroquinone, 0.5-1 mM chloranil, 2-10 mM L-cysteine, and 0.1-0.5 mM tris(2-carboxyethyl)phosphine have the effect on TETase Facilitation works best.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com