Method and system for rapid biomolecular recognition of amino acids and protein sequencing
a biomolecular recognition and amino acid technology, applied in the field of methods and systems for rapid biomolecular recognition of amino acids and protein sequencing, can solve the problems of complex methods, labor-intensive amino acid analysis and protein end group analysis methods, and insensitivity, and achieve the effect of low cost and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
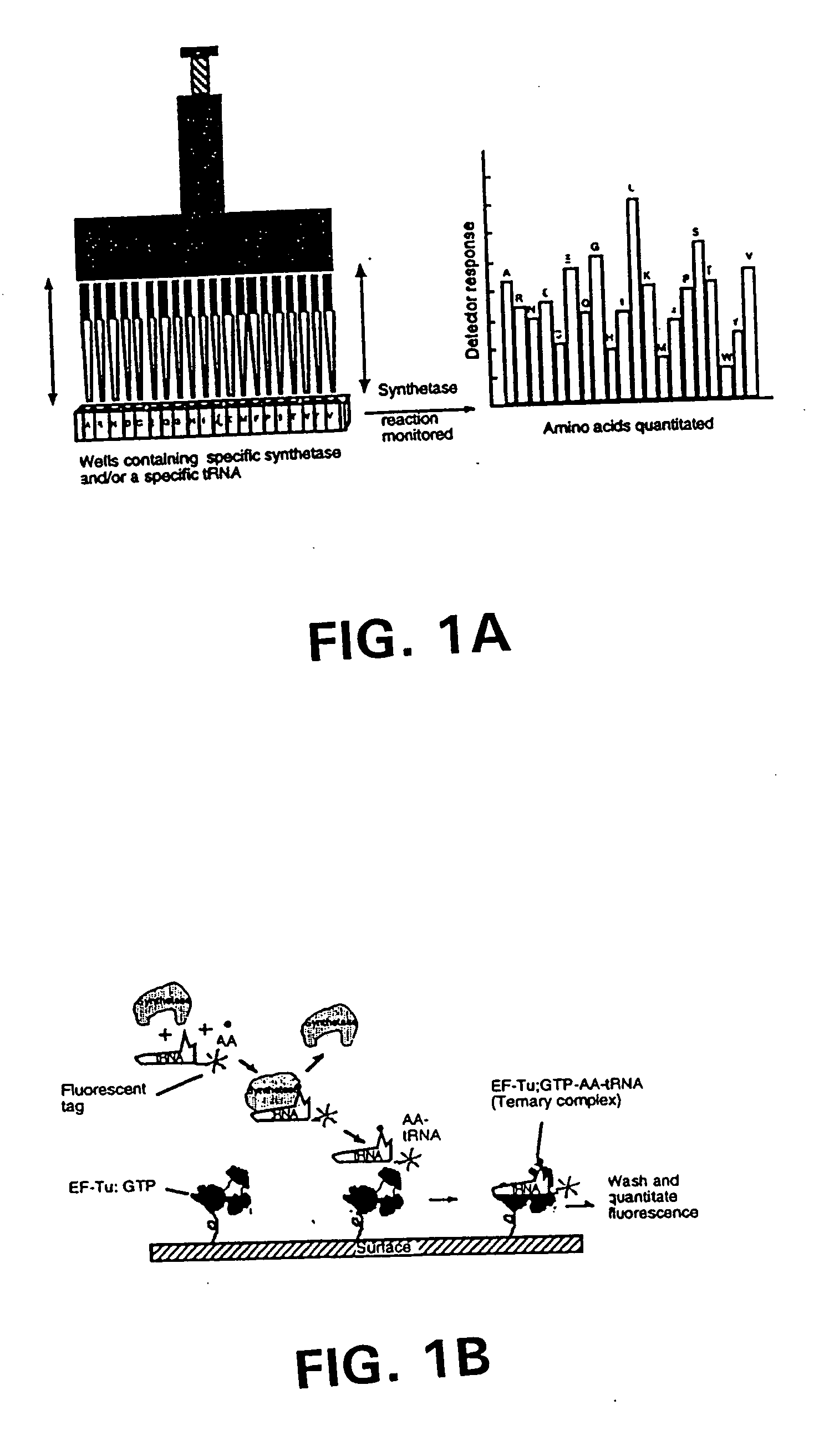
[0279] Example 1 illustrates the use of an array to simultaneously and quantitatively detect each of the individual primary amino acids in a sample. As shown in FIG. 1A, the 20 aminoacyl-tRNA synthetases and / or a cognate tRNA specific for a different amino acid can be arranged in array format. By dispensing each synthetase into a different well at a known position, the amino acid-specific reactions catalyzed by these enzymes can be used to identify and quantitate their cognate amino acids. Each well thereby would signal only the amino acid cognate to the synthetase and / or tRNA present in that well. The single letter codes for the amino acids are used in this figure to represent the synthetase or tRNA cognate for the corresponding amino acid. In this embodiment, as shown in FIG. 1A, the reactions catalyzed by the synthetases are carried out in parallel and are monitored in all the wells simultaneously by use of a plate reader. The presence and amounts of the various individual amino ...
example 2
[0281] Example 2 describes several different embodiments wherein an elongation factor is used in the detection of a ternary complex.
Immobilization of the EF-Tu:GTP with Labeling of the tRNA.
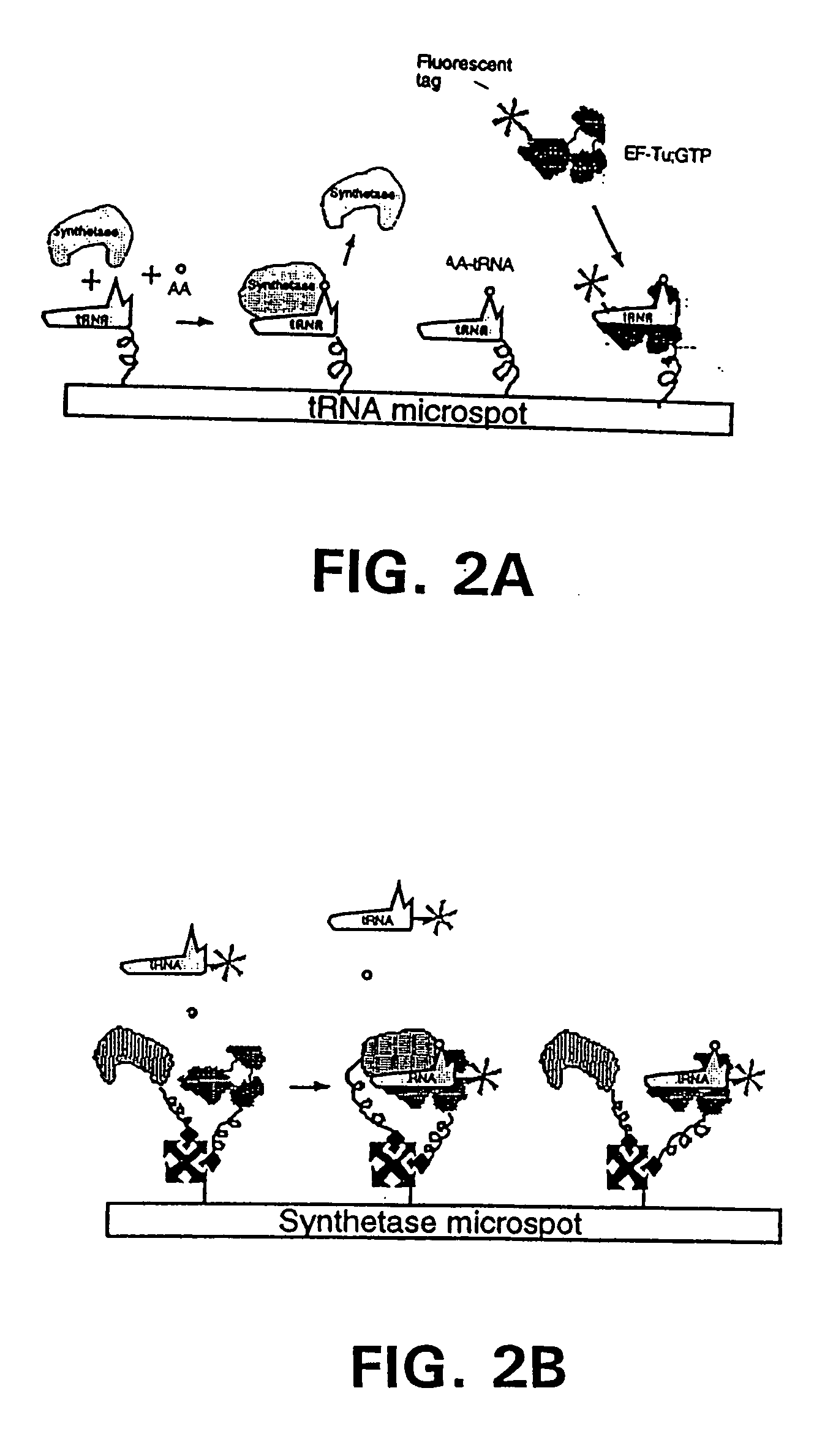
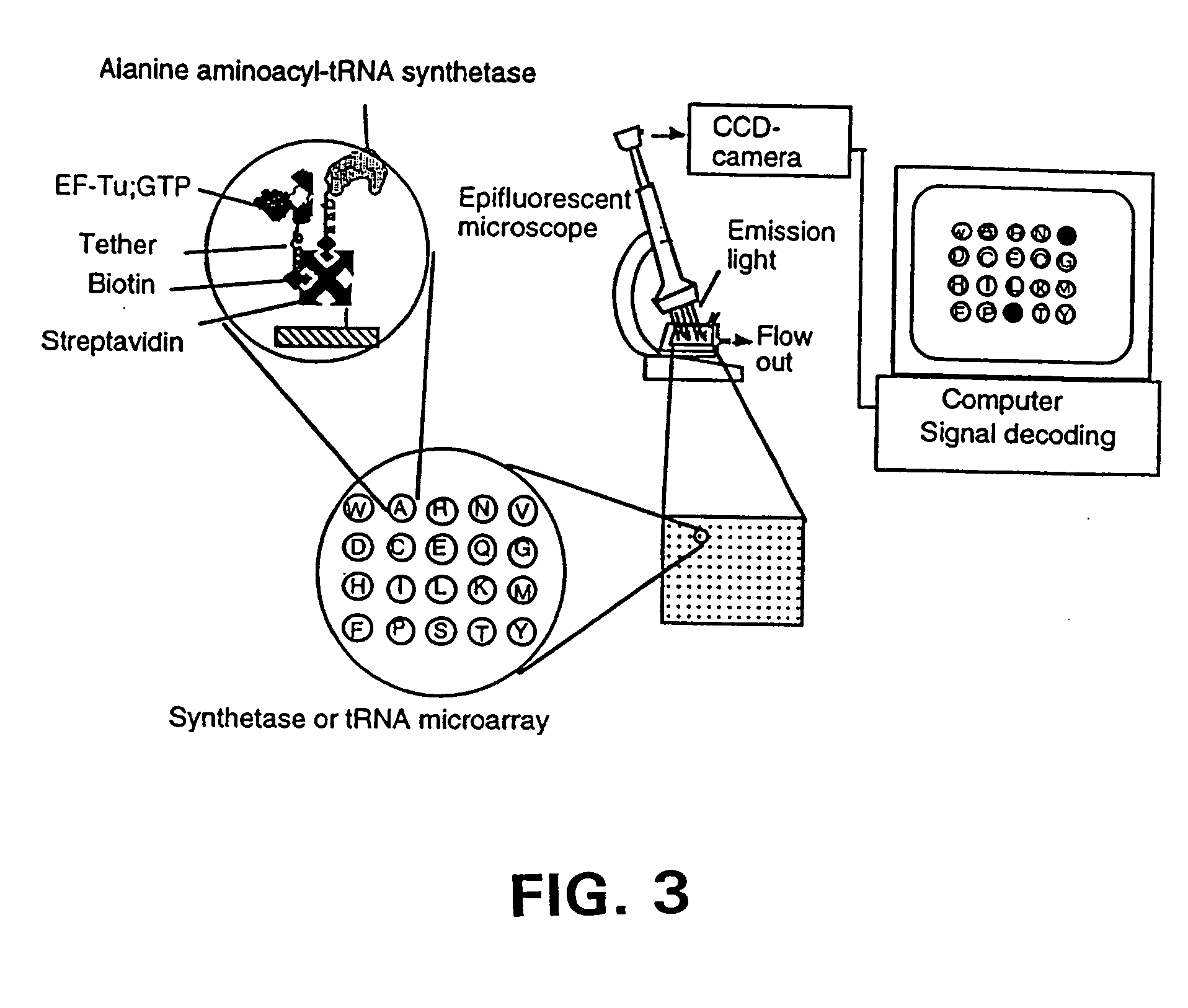
[0282] In a first embodiment as shown in FIG. 1B, EF-Tu:GTP is immobilized to the surface of a reaction site (e.g., the bottom of a microtiter wells). The shown site may be one of many sites providing aminoacyl tRNA synthetase reactions and having immobilized EF-Tu:GTP. In a preferred embodiment, there are such sites for each of the 20 primary amino acids containing a different one of the 20 synthetases in aminoacylation buffer and its labeled cognate tRNA. As shown in FIG. 1B, the tRNA is attached to a label, which in this case is a fluorescent label. After the amino acid mixture or sample is added to the reaction site, the aminoacyl tRNA synthetase catalyzes the formation of the correspondingly labeled aminoacyl tRNA. The labeled aminoacyl tRNA binds to the immobilized EF-Tu:GTP to form a la...
example 3
[0285] The reaction conditions and buffers suitable for binding of elongation factors to aminoacyl tRNAs are well known to one of ordinary skill in the art. Examples of suitable reaction buffers for monitoring the binding of EF-Tu:GTP to the AA−tRNAs include but are not limited to: [0286] 1. 50 mM Tris-HCl, pH 7.5, 50 mM NH4Cl, 50M KCl, 10 mM MgCl2, 1 mM GTP, 5 mM beta-mercaptoethanol, 1 mM ATP, 23°deg C. [0287] 2. 50 mM Tris-HCl, pH 7.5, 25 mM KCl, 5 mM MgCl, 2 mM beta-mercaptoethanol, 10 mM ATP, 10 mM phosphoenolpyruvate, 120 ug / ml pyruvate kinase, 1 mM GTP
[0288] The binding of the AA−tRNAs to the EF-Tu:GTP may be performed at room temperature when using the elongation factor from T. thermophilus, for example, taking advantage of the thermal stability of this protein. In brief, the AA−tRNAs are reacted with the EF-Tu:GTP in reaction buffer. The EF-Tu:GTP is optionally bound to a surface.
[0289] In cases where one molecule is immobilized and used to “capture” another molecule a wa...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com