Raw maltogenic amylase production bacterial strain
A technology of maltose amylase and bacteria, which is applied in the field of genetic engineering, can solve problems such as the reduction of the final yield of maltose and the reduction of product purity, and achieve the effects of improved temperature stability, high catalytic efficiency and low by-products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
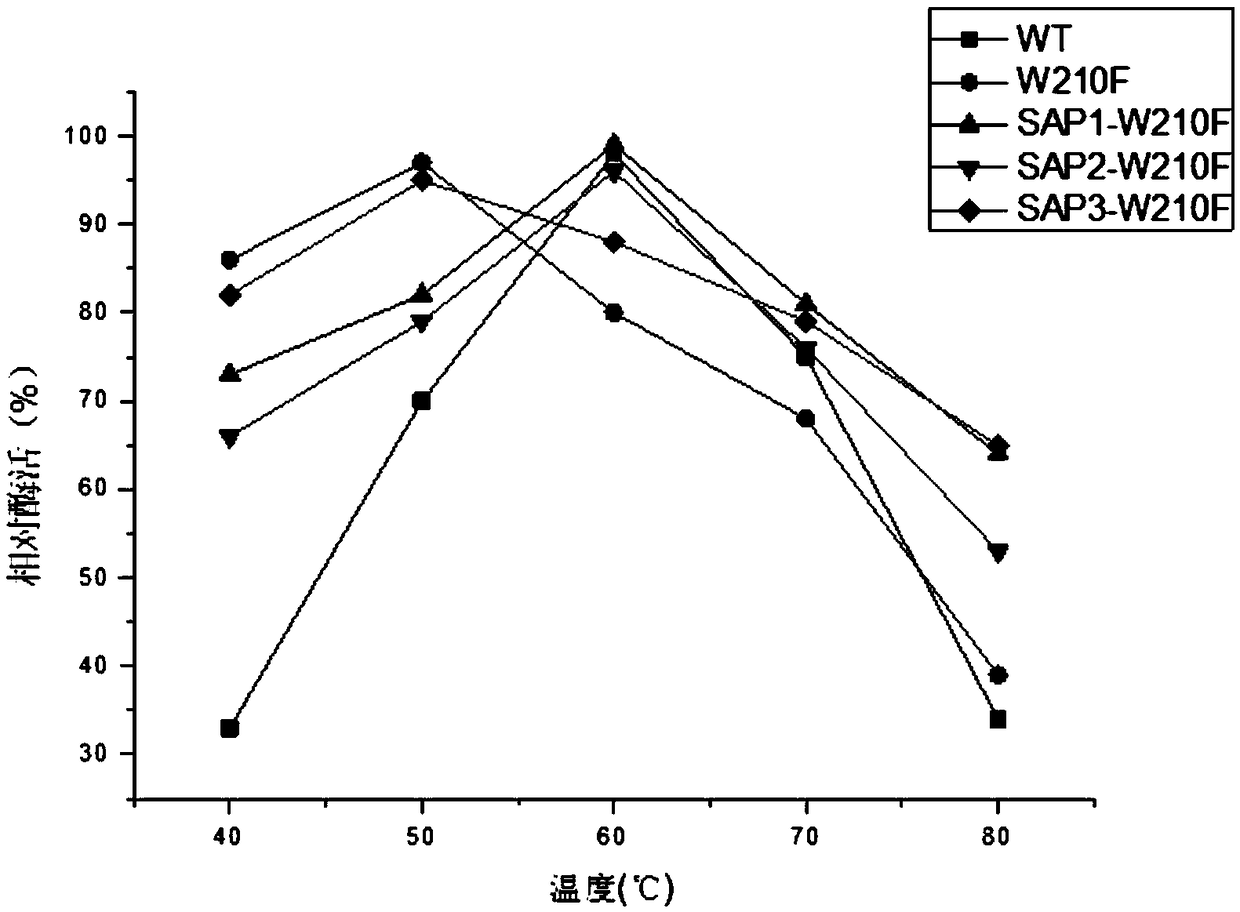
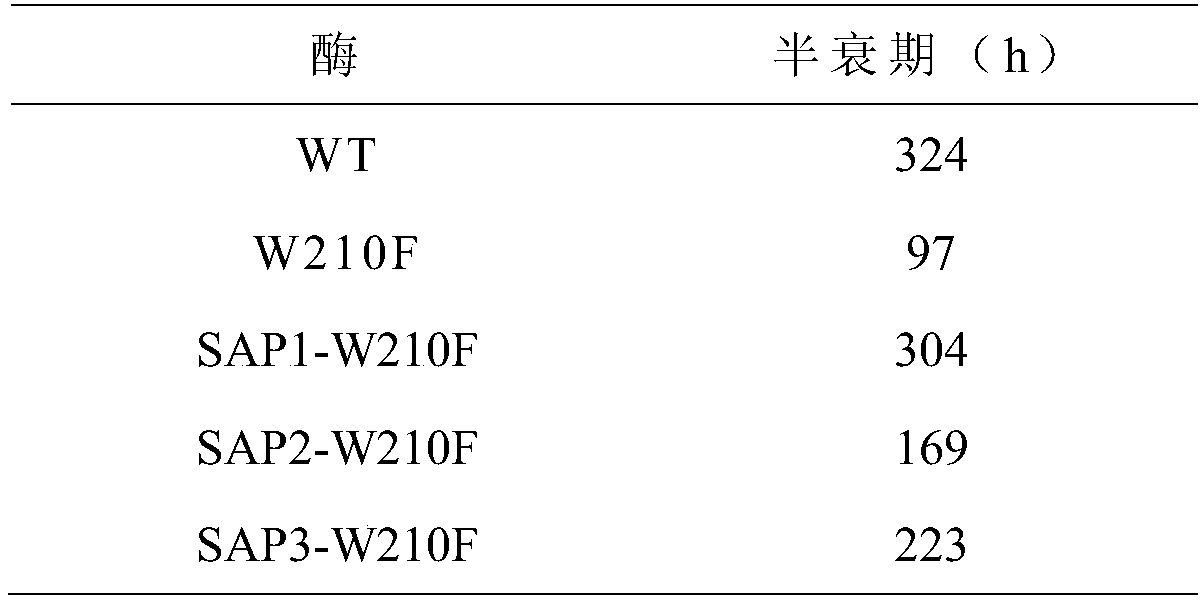
Image
Examples
Embodiment 1
[0021] Example 1: Preparation of wild raw maltogenic amylase.
[0022] (1) Construction of Maltogenic Amylase Recombinant Bacteria
[0023] According to the amyM amino acid sequence on NCBI (NCBI number: AAA22233.1), the sequence was codon-optimized, and the gene sequence amyM of maltogenic amylase was synthesized by chemical total synthesis. The plasmid used to construct the expression vector in E. coli is pET24a(+). The pET24a(+) plasmid and the plasmid with the amyM gene were subjected to NcoI and HindIII double enzyme digestion respectively. After the digestion products were recovered by gel, they were ligated with T4 ligase overnight, and the ligated products were transformed into Escherichia coli JM109 competent cells, and the transformed products Spread on LB plates containing 100mg / L kanamycin, culture overnight at 37°C, pick 2 single colonies on the plate, insert them into LB liquid medium, extract plasmids for verification after 8 hours, the result is correct, and e...
Embodiment 2
[0027] Embodiment 2: Preparation of raw maltogenic amylase mutants
[0028] (1) Replace tryptophan (Trp) at position 210 in maltogenic amylase with phenylalanine (Phe), denoted as W210F.
[0029] The site-directed mutagenesis primers for introducing the W210F mutation are:
[0030] Forward primer: 5'-TGACATCTCTAAC TTC GACGACCGTTACGA-3' (the underline is the mutated base)
[0031] Reverse primer: 5'-TCGTAACGGTCGTC GAA GTTAGAGATGTCA-3' (the underline is the mutated base)
[0032] The pET24a-amyM plasmid was used as a template for PCR reaction. All reactions were carried out in a 50 μL system, and the reaction conditions were: pre-denaturation at 94°C for 4 min; followed by 30 cycles (94°C for 10 s, 50°C for 10 s, 72°C for 7 min and 20 s); extension at 72°C for 10 min; and finally incubation at 4°C. The PCR products were digested with DpnⅠ (Fermentas Company), and transformed into Escherichia coli JM109 competent cells, respectively, and the transformed products were sprea...
Embodiment 3
[0040] Embodiment 3: Enzyme activity analysis of raw maltose amylase
[0041] (1) Definition of enzyme activity unit
[0042] When using the 3,5-dinitrosalicylic acid method (DNS method) to measure the activity of maltogenic amylase, the amount of enzyme needed to catalyze the production of 1 μmol of reducing sugar per minute is taken as an activity unit.
[0043] (2) Enzyme Activity Determination Steps
[0044] Preheating: Take 2mL of 0.5% soluble starch solution (50mM pH5.5 citric acid buffer) in a test tube and place it in a 60°C water bath to preheat for 10min.
[0045] Reaction: Add 0.1mL sample enzyme solution, oscillate evenly, accurately time 10min, add 3mL DNS, oscillate evenly, put into ice water to terminate the reaction, boil in a boiling water bath for 7min. cool down.
[0046] Measurement: add distilled water to the above reaction system and adjust the volume to 15mL, and mix well. The absorbance was measured at a wavelength of 540nm and the enzyme activity w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com